Journal:Acta Cryst D:S2059798325007089
From Proteopedia
(Difference between revisions)

| Line 10: | Line 10: | ||
These findings show that cryo-EM can capture not only the intended targets but also unexpected complexes that are well resolved and may have physiological importance. While only partial densities of the FtsH AAA+ domain were visible and no stable FtsH–YidC–HflKC complex could be reconstructed, the high-quality ArnA and AcrB structures provide fresh insights into bacterial survival strategies, from antibiotic resistance to drug efflux. More broadly, this study illustrates how structural biology can reveal unplanned discoveries that enrich our understanding of cell biology and the challenges of protein purification. | These findings show that cryo-EM can capture not only the intended targets but also unexpected complexes that are well resolved and may have physiological importance. While only partial densities of the FtsH AAA+ domain were visible and no stable FtsH–YidC–HflKC complex could be reconstructed, the high-quality ArnA and AcrB structures provide fresh insights into bacterial survival strategies, from antibiotic resistance to drug efflux. More broadly, this study illustrates how structural biology can reveal unplanned discoveries that enrich our understanding of cell biology and the challenges of protein purification. | ||
| + | |||
| + | |||
| + | {| class="wikitable" style="margin:auto" | ||
| + | |- | ||
| + | |- | ||
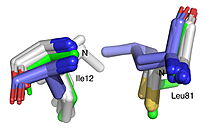
| + | | [[Image:022_Fig2.jpg label.jpg|thumb|left|250px|Best fit overlay of the Cɑ positions of SABP2 structures (three structures, light blue carbons) and HbHNL structures (eighteen structures, white carbons) onto the structure of | ||
| + | HNL6V (green sticks). The catalytic atoms of the catalytic triad of this α/β-hydrolase fold, in HNL6V (Oɣ of S80, Nε2 of H235, Oδ2 of D207) overlay more closely with the corresponding atoms in the HbHNL structures (S80, H235, A207) than with the corresponding atoms in the SABP2 structures (S81, H238, D210).]] [[Image:019_Fig_3b.oxyanion_label.jpg|thumb|right|210px|Best fit overlay of the Cɑ positions of SABP2 structures (three structures, light blue carbons) and HbHNL structures (eighteen structures, white carbons) onto the structure of HNL6V (green sticks). The oxyanion hole amide nitrogen atoms of I12 and L81 in HNL6V overlay more closely with the corresponding atoms in HbHNL (I12, C81) than with the corresponding atoms in SABP2 (A13 and L82).]] | ||
| + | |} | ||
| + | |||
<b>References</b><br> | <b>References</b><br> | ||
Revision as of 07:23, 18 September 2025
| |||||||||||
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.