Sandbox Home
From Proteopedia
| Line 1: | Line 1: | ||
| - | + | table id="tableColumnsMainPage" style="width:100%;border:2px solid #ddd;border-collapse: collapse;table-layout: fixed; "> | |
| + | <tr><td colspan='3' style="background:#F5F5FC;border:1px solid #ddd;"> | ||
| + | <div style="top:+0.2em; font-size:1.2em; padding:5px 5px 5px 10px; float:right;">'''''ISSN 2310-6301'''''</div> | ||
| - | + | <span style="border:none; margin:0; padding:0.3em; color:#000; font-style: italic; font-size: 1.4em;"> | |
| - | + | <b>As life is more than 2D</b>, Proteopedia helps to bridge the gap between 3D structure & function of biomacromolecules | |
| - | + | </span> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | <span style="border:none; margin:0; padding:0.3em; color:#000; font-style: italic; font-size: 1.1em;max-width:80%;display:block;"> | |
| - | + | <b>Proteopedia</b> presents this information in a user-friendly way as a '''collaborative & free 3D-encyclopedia of proteins & other biomolecules.''' | |
| - | + | </span> | |
| - | <span style="display:block; margin:0; padding:0.3em; color:#000; font-style:italic; font-size:1.1em; max-width:80%;"> | ||
| - | <b>Proteopedia</b> presents this information in a user-friendly way as a <b>collaborative & free 3D-encyclopedia of proteins & other biomolecules.</b> | ||
| - | </span> | ||
| - | + | </td></tr> | |
| - | + | ||
| - | + | ||
| - | + | <tr> | |
| - | + | <th style="padding: 10px;background-color: #33ff7b">Selected Research Pages</th> | |
| - | + | <th style="padding: 10px;background-color: #f1b840">In Journals</th> | |
| - | + | <th style="padding: 10px;background-color: #79baff">Education</th> | |
| - | + | </tr> | |
| - | + | ||
| - | + | <tr valign='top'> | |
| - | + | <td style="padding: 5px;"> {{Proteopedia:Featured SEL/{{#expr: {{#time:U}} mod {{Proteopedia:Number of SEL articles}}}}}}</td> | |
| - | + | <td style="padding: 5px;"> {{Proteopedia:Featured JRN/{{#expr: {{#time:U}} mod {{Proteopedia:Number of JRN articles}}}}}}</td> | |
| - | + | <td style="padding: 5px;"> {{Proteopedia:Featured EDU/{{#expr: {{#time:U}} mod {{Proteopedia:Number of EDU articles}}}}}}</td> | |
| - | + | </tr> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | <tr style="font-size: 1.2em; text-align: center;"> | |
| - | + | <td style="padding: 10px;background-color: #33ff7b"></td> | |
| - | + | <td style="padding: 10px;background-color: #f1b840"></td> | |
| - | + | <td style="padding: 10px;background-color: #79baff"></td> | |
| - | + | </tr> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | <!-- RIGHT COLUMN --> | ||
| - | <td style="padding:10px; vertical-align:top;"> | ||
| - | <div> | ||
| - | <p>[[Teaching strategies using Proteopedia]]</p> | ||
| - | <p>[[Teaching_Scenes%2C_Tutorials%2C_and_Educators%27_Pages|Examples of pages for teaching]]</p> | ||
| - | <p>[[Help:Contents#For_authors:_contributing_content|How to add content to Proteopedia]]</p> | ||
| - | </div> | ||
| - | <div style="margin-top:10px;"> | ||
| - | {{Proteopedia:Featured EDU/{{#expr: {{#time:U}} mod {{Proteopedia:Number of EDU articles}}}}}} | ||
| - | </div> | ||
| - | </td> | ||
| - | </tr> | ||
| - | + | <tr style="font-size: 1.0em; text-align: center;"> | |
| - | + | ||
| - | + | <td style="padding: 10px;> | |
| - | + | <p>[[Help:Contents#For_authors:_contributing_content|How to add content to Proteopedia]]</p> | |
| - | + | <p>[[Proteopedia:Video_Guide|Video Guides]]</p> | |
| - | + | <p>[[Who knows]] ...</p> | |
| - | + | </td> | |
| - | + | ||
| - | + | <td style="padding: 10px;> | |
| - | + | <p>[[I3DC|About Interactive 3D Complements - '''I3DCs''']]</p> | |
| - | + | <p>[[Proteopedia:I3DC|List of I3DCs]]</p> | |
| - | + | <p>[[How to get an I3DC for your paper]]</p> | |
| - | + | ||
| - | + | </td> | |
| - | + | ||
| + | <td style="padding: 10px;> | ||
| + | <p>[[Teaching strategies using Proteopedia]]</p> | ||
| + | <p>[[Teaching_Scenes%2C_Tutorials%2C_and_Educators%27_Pages|Examples of pages for teaching]]</p> | ||
| + | <p>[[Help:Contents#For_authors:_contributing_content|How to add content to Proteopedia]]</p> | ||
| + | </td> | ||
| + | |||
| + | </tr> | ||
| + | |||
| + | <tr><td colspan='3' > | ||
| + | <table width='100%' style="padding: 10px; background-color: #d7d8f9; font-size: 1.5em;"><tr> | ||
| + | <td>[[Proteopedia:About|About]]</td> | ||
| + | <td>[[Special:Contact|Contact]]</td> | ||
| + | <td>[[Template:MainPageNews|Hot News]]</td> | ||
| + | <td>[[Proteopedia:Table of Contents|Table of Contents]]</td> | ||
| + | <td>[[Proteopedia:Structure Index|Structure Index]]</td> | ||
| + | <td>[[Help:Contents|Help]]</td> | ||
| + | </tr></table> | ||
| + | </td></tr> | ||
</table> | </table> | ||
Revision as of 16:09, 30 September 2025
table id="tableColumnsMainPage" style="width:100%;border:2px solid #ddd;border-collapse: collapse;table-layout: fixed; "> <tr><td colspan='3' style="background:#F5F5FC;border:1px solid #ddd;">
As life is more than 2D, Proteopedia helps to bridge the gap between 3D structure & function of biomacromolecules
Proteopedia presents this information in a user-friendly way as a collaborative & free 3D-encyclopedia of proteins & other biomolecules.
</td></tr>
<tr> <th style="padding: 10px;background-color: #33ff7b">Selected Research Pages</th> <th style="padding: 10px;background-color: #f1b840">In Journals</th> <th style="padding: 10px;background-color: #79baff">Education</th> </tr>
<tr valign='top'>
<td style="padding: 5px;">BREAKTHROUGH in protein structure prediction!
by Eric Martz |
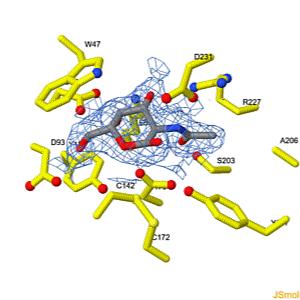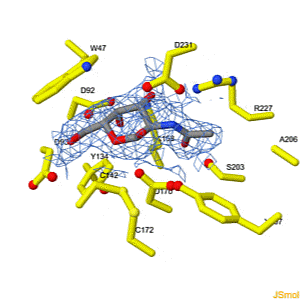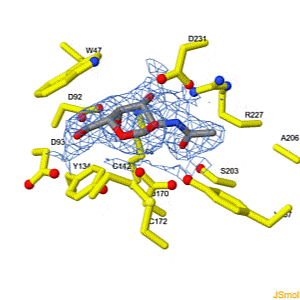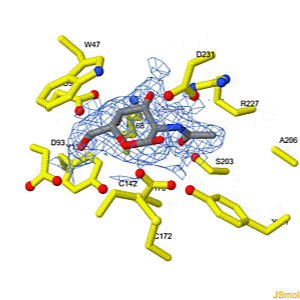
Interconversion of the specificities of human lysosomal enzymes associated with Fabry and Schindler diseases.
IB Tomasic, MC Metcalf, AI Guce, NE Clark, SC Garman. J. Biol. Chem. 2010 doi: 10.1074/jbc.M110.118588 >>> Visit this I3DC complement >>> |
Make Your Own Electrostatic Potential Maps
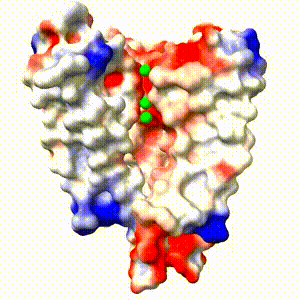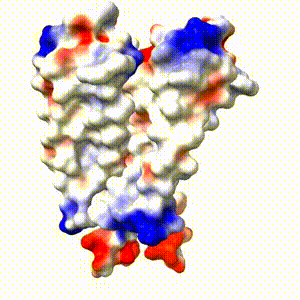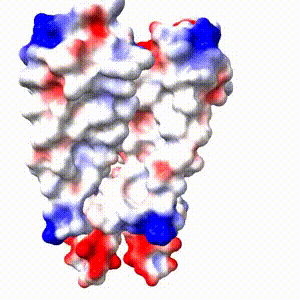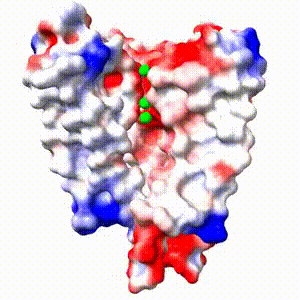
Positive (+) and Negative (-) charges on the surface of a protein molecule play crucial roles in its interactions with other molecules, and hence in its functions. Electrostatic potential maps coloring the surface of a protein molecule are a popular way to visualize the distribution of surface charges. Easy to use free software is available to to create these surface maps. Above is an integral membrane potassium channel protein. One of its 4 identical chains is removed so you can see the Negative (-) protein surface contacting the 3 K+ ions. |
</tr>
<tr style="font-size: 1.2em; text-align: center;"> <td style="padding: 10px;background-color: #33ff7b"></td> <td style="padding: 10px;background-color: #f1b840"></td> <td style="padding: 10px;background-color: #79baff"></td> </tr>
<tr style="font-size: 1.0em; text-align: center;">
<td style="padding: 10px;>
How to add content to Proteopedia
Who knows ...
</td>
<td style="padding: 10px;>
About Interactive 3D Complements - I3DCs
How to get an I3DC for your paper
</td>
<td style="padding: 10px;>
Teaching strategies using Proteopedia
Examples of pages for teaching
How to add content to Proteopedia
</td>
</tr>
<tr><td colspan='3' >
| About | Contact | Hot News | Table of Contents | Structure Index | Help |
</td></tr> </table>