Kaushki Sharma- BI3323
From Proteopedia
(Difference between revisions)
| Line 42: | Line 42: | ||
Funding Organization(s): National Research Foundation (NRF, Korea) | Funding Organization(s): National Research Foundation (NRF, Korea) | ||
| - | |||
'''Experimental Data Snapshot''' | '''Experimental Data Snapshot''' | ||
| Line 119: | Line 118: | ||
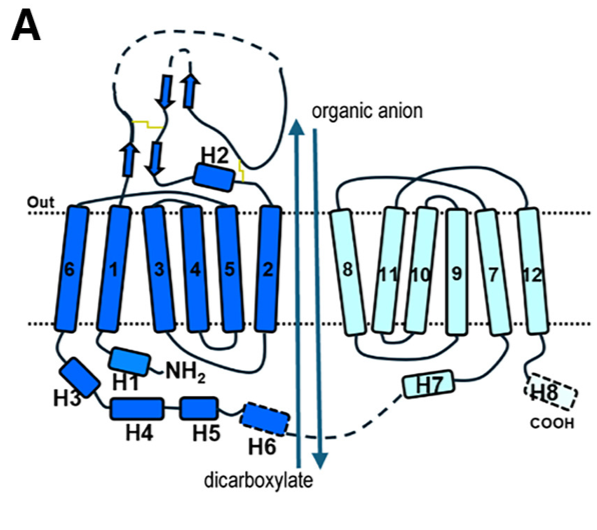
*Olmesartan occupies Site 3 of the binding pocket and is located within 5A˚ distance of residues of TM1, TM4, TM5, TM7, TM10, and TM11, namely N35, M207, G227, Y230, W346, Y353, Y354, F438, F442, S462, and R466. | *Olmesartan occupies Site 3 of the binding pocket and is located within 5A˚ distance of residues of TM1, TM4, TM5, TM7, TM10, and TM11, namely N35, M207, G227, Y230, W346, Y353, Y354, F438, F442, S462, and R466. | ||
| - | |||
| - | |||
| - | |||
===Mechanism of OAT1 inhibition by probenecid=== | ===Mechanism of OAT1 inhibition by probenecid=== | ||
| Line 141: | Line 137: | ||
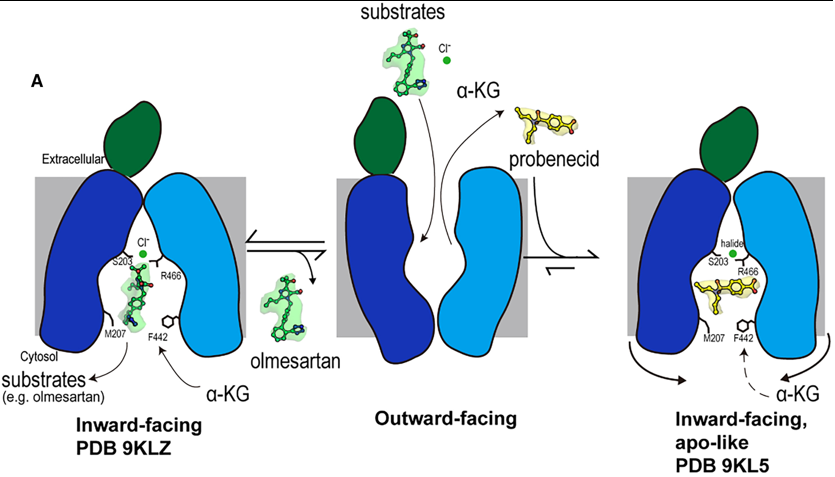
By constricting the cytoplasmic access routes, probenecid does not just compete for the substrate-binding site; it stabilizes the transporter in an apo-like, inward-facing conformation that is inaccessible to cytosolic substrates. This prevents the entry of new substrates and likely traps the transporter in this non-functional state, effectively "locking" it and preventing the conformational changes necessary for the transport cycle. | By constricting the cytoplasmic access routes, probenecid does not just compete for the substrate-binding site; it stabilizes the transporter in an apo-like, inward-facing conformation that is inaccessible to cytosolic substrates. This prevents the entry of new substrates and likely traps the transporter in this non-functional state, effectively "locking" it and preventing the conformational changes necessary for the transport cycle. | ||
| - | |||
| - | |||
===Mechanistic Insights into hOAT1 Function and Inhibition=== | ===Mechanistic Insights into hOAT1 Function and Inhibition=== | ||
| Line 151: | Line 145: | ||
conformation change for inhibition (apo-like conformation).]] | conformation change for inhibition (apo-like conformation).]] | ||
| - | ''' | + | ''' A Dual-Mechanism for Potent Inhibition by Probenecid''' |
The study reveals that the classic inhibitor probenecid employs a sophisticated, dual-mechanism to arrest OAT1 function, moving beyond simple competition. | The study reveals that the classic inhibitor probenecid employs a sophisticated, dual-mechanism to arrest OAT1 function, moving beyond simple competition. | ||
| Line 159: | Line 153: | ||
'''Conformational Arrest:''' More significantly, probenecid binding induces subtle conformational changes in the cytoplasmic ends of transmembrane helices (TM5, TM8, TM10, TM11). This leads to a constriction of the cytosolic opening, completely blocking one access path (Path B) and narrowing the other (Path A). This physically prevents substrates from entering or exiting the binding site from the cytoplasm, effectively "locking" the transporter in an inactive, inward-facing state. This mechanism is reminiscent of inhibition seen in other transporters like hURAT1, suggesting it may be a general strategy for effective transport arrest. | '''Conformational Arrest:''' More significantly, probenecid binding induces subtle conformational changes in the cytoplasmic ends of transmembrane helices (TM5, TM8, TM10, TM11). This leads to a constriction of the cytosolic opening, completely blocking one access path (Path B) and narrowing the other (Path A). This physically prevents substrates from entering or exiting the binding site from the cytoplasm, effectively "locking" the transporter in an inactive, inward-facing state. This mechanism is reminiscent of inhibition seen in other transporters like hURAT1, suggesting it may be a general strategy for effective transport arrest. | ||
| - | + | ===Conclusion=== | |
| - | + | rOAT1 structures with probenecid have been reported previously, <ref>Parker, J.L., Kato, T., Kuteyi, G., Sitsel, O., and Newstead, S. (2023). | |
| - | + | Molecular basis for selective uptake and elimination of organic anions in | |
| - | + | the kidney by OAT1. Nat. Struct. Mol. Biol. 30, 1786–1793. https://doi. | |
| - | + | org/10.1038/s41594-023-01039-y.</ref> and our hOAT1 structures align with findings for rOAT1 and provide new insights into the mechanism by which probenecid inhibits transport activity. Additionally, this study reveals the structure of hOAT1 with olmesartan, offering mechanistic insights into species-specific differences in OAT1 transport of specific substrates. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
==Notes & References== | ==Notes & References== | ||
<references /> | <references /> | ||
Revision as of 17:49, 30 November 2025
Interactive 3D Complement in Proteopedia
|
| |
|
Cryo-EM structures of human OAT1 reveal drug binding and inhibition mechanisms[1]. | |
|
Cell Volume 33, Issue 11, P1856-1866.E5, November 06, 2025 |
Structure Tour
| |||||||||||