Sandbox Aryan 20221057 BI3323-Aug2025
From Proteopedia
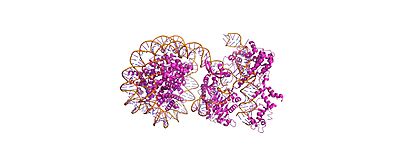
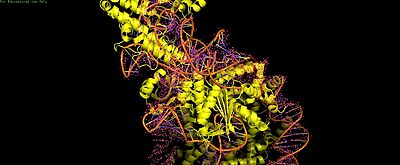
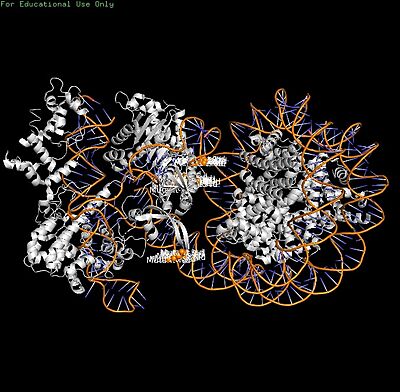
(== Summary == PDB **8YNY** (4.52 Å cryo-EM) captures Cas9-sgRNA post-cleavage binding to nucleosome linker DNA (PAM1).[web:2] <Jmol width=100% height=500px>load =8YNY; cartoon on; color chain; spin y) |
Revision as of 18:31, 30 November 2025
Contents |
Structure Overview
Cas9-sgRNA ribonucleoprotein targets nucleosome **linker DNA** (PAM1/PAM28) and **entry-exit regions** (SHL6), avoiding tightly wrapped **core DNA** (SHL0-5). Native-PAGE on Widom 601 nucleosomes confirmed preferential cleavage at DNA ends where transient unwrapping occurs.[attached_file:1]
The post-cleavage complex shows HNH/REC2 domains disordered, bridge helix absent, and target/non-target DNA strands cleaved—consistent with binary biochemical data.[web:2]
PI Domain Interactions
Cas9's **PI domain** (residues ~1100-1368) makes multiple contacts: - **Histone tails**: Weak electrostatic interaction (non-essential for binding) - **PI edge (K1155)**: Lysine stabilizes post-cleavage complex via DNA phosphate backbone - **Core DNA loops (H1264/R1298/K1300)**: Nonspecific binding inhibits cleavage
- Mutagenesis validation**: H1264A/R1298Q/K1300A mutants increase nucleosome binding AND cleavage efficiency both in vitro and rice callus genome editing.[attached_file:1]
Dual Inhibition Mechanism
1. **Access barrier**: Nucleosome DNA ends inflexible (SHL0-5), blocking Cas9 binding 2. **Motion restriction**: PI-core DNA trapping limits HNH/RuvC domain movements for cleavage
- Entry/exit asymmetry** from Widom601 sequence flexibility explains variable editing across chromatin contexts.[web:14]
Implications
Reveals Cas9's eukaryotic adaptation strategy and identifies **chromatin-optimized variants** for improved genome editing tools.[web:121]
BI3323-Aug2025