1bob
From Proteopedia
(New page: 200px<br /><applet load="1bob" size="450" color="white" frame="true" align="right" spinBox="true" caption="1bob, resolution 2.3Å" /> '''HISTONE ACETYLTRANSFE...) |
|||
| Line 1: | Line 1: | ||
| - | [[Image:1bob.jpg|left|200px]]<br /><applet load="1bob" size=" | + | [[Image:1bob.jpg|left|200px]]<br /><applet load="1bob" size="350" color="white" frame="true" align="right" spinBox="true" |
caption="1bob, resolution 2.3Å" /> | caption="1bob, resolution 2.3Å" /> | ||
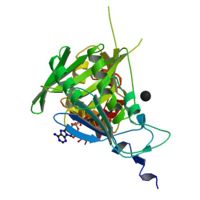
'''HISTONE ACETYLTRANSFERASE HAT1 FROM SACCHAROMYCES CEREVISIAE IN COMPLEX WITH ACETYL COENZYME A'''<br /> | '''HISTONE ACETYLTRANSFERASE HAT1 FROM SACCHAROMYCES CEREVISIAE IN COMPLEX WITH ACETYL COENZYME A'''<br /> | ||
==Overview== | ==Overview== | ||
| - | We have solved the crystal structure of the yeast histone | + | We have solved the crystal structure of the yeast histone acetyltransferase Hat1-acetyl coenzyme A (AcCoA) complex at 2.3 A resolution. Hat1 has an elongated, curved structure, and the AcCoA molecule is bound in a cleft on the concave surface of the protein, marking the active site of the enzyme. A channel of variable width and depth that runs across the protein is probably the binding site for the histone substrate. A model for histone H4 binding by Hat1 is discussed in terms of possible sources of specific lysine recognition by the enzyme. The structure of Hat1 provides a model for the structures of the catalytic domains of a protein superfamily that includes other histone acetyltransferases such as Gcn5 and CBP. |
==About this Structure== | ==About this Structure== | ||
| - | 1BOB is a [http://en.wikipedia.org/wiki/Single_protein Single protein] structure of sequence from [http://en.wikipedia.org/wiki/Saccharomyces_cerevisiae Saccharomyces cerevisiae] with CA and ACO as [http://en.wikipedia.org/wiki/ligands ligands]. Active as [http://en.wikipedia.org/wiki/Histone_acetyltransferase Histone acetyltransferase], with EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=2.3.1.48 2.3.1.48] Full crystallographic information is available from [http:// | + | 1BOB is a [http://en.wikipedia.org/wiki/Single_protein Single protein] structure of sequence from [http://en.wikipedia.org/wiki/Saccharomyces_cerevisiae Saccharomyces cerevisiae] with <scene name='pdbligand=CA:'>CA</scene> and <scene name='pdbligand=ACO:'>ACO</scene> as [http://en.wikipedia.org/wiki/ligands ligands]. Active as [http://en.wikipedia.org/wiki/Histone_acetyltransferase Histone acetyltransferase], with EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=2.3.1.48 2.3.1.48] Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1BOB OCA]. |
==Reference== | ==Reference== | ||
| Line 14: | Line 14: | ||
[[Category: Saccharomyces cerevisiae]] | [[Category: Saccharomyces cerevisiae]] | ||
[[Category: Single protein]] | [[Category: Single protein]] | ||
| - | [[Category: Dutnall, R | + | [[Category: Dutnall, R N.]] |
[[Category: Ramakrishnan, V.]] | [[Category: Ramakrishnan, V.]] | ||
[[Category: Sternglanz, R.]] | [[Category: Sternglanz, R.]] | ||
| - | [[Category: Tafrov, S | + | [[Category: Tafrov, S T.]] |
[[Category: ACO]] | [[Category: ACO]] | ||
[[Category: CA]] | [[Category: CA]] | ||
| Line 24: | Line 24: | ||
[[Category: histone modification]] | [[Category: histone modification]] | ||
| - | ''Page seeded by [http:// | + | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Thu Feb 21 11:57:25 2008'' |
Revision as of 09:57, 21 February 2008
|
HISTONE ACETYLTRANSFERASE HAT1 FROM SACCHAROMYCES CEREVISIAE IN COMPLEX WITH ACETYL COENZYME A
Overview
We have solved the crystal structure of the yeast histone acetyltransferase Hat1-acetyl coenzyme A (AcCoA) complex at 2.3 A resolution. Hat1 has an elongated, curved structure, and the AcCoA molecule is bound in a cleft on the concave surface of the protein, marking the active site of the enzyme. A channel of variable width and depth that runs across the protein is probably the binding site for the histone substrate. A model for histone H4 binding by Hat1 is discussed in terms of possible sources of specific lysine recognition by the enzyme. The structure of Hat1 provides a model for the structures of the catalytic domains of a protein superfamily that includes other histone acetyltransferases such as Gcn5 and CBP.
About this Structure
1BOB is a Single protein structure of sequence from Saccharomyces cerevisiae with and as ligands. Active as Histone acetyltransferase, with EC number 2.3.1.48 Full crystallographic information is available from OCA.
Reference
Structure of the histone acetyltransferase Hat1: a paradigm for the GCN5-related N-acetyltransferase superfamily., Dutnall RN, Tafrov ST, Sternglanz R, Ramakrishnan V, Cell. 1998 Aug 21;94(4):427-38. PMID:9727486
Page seeded by OCA on Thu Feb 21 11:57:25 2008