1fiq
From Proteopedia
(New page: 200px<br /><applet load="1fiq" size="450" color="white" frame="true" align="right" spinBox="true" caption="1fiq, resolution 2.50Å" /> '''CRYSTAL STRUCTURE OF...) |
|||
| Line 1: | Line 1: | ||
| - | [[Image:1fiq.gif|left|200px]]<br /><applet load="1fiq" size=" | + | [[Image:1fiq.gif|left|200px]]<br /><applet load="1fiq" size="350" color="white" frame="true" align="right" spinBox="true" |
caption="1fiq, resolution 2.50Å" /> | caption="1fiq, resolution 2.50Å" /> | ||
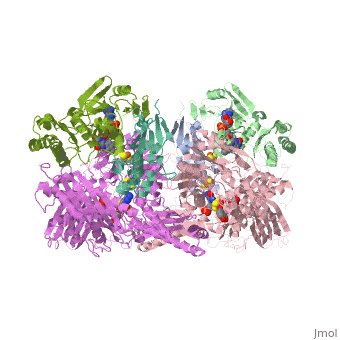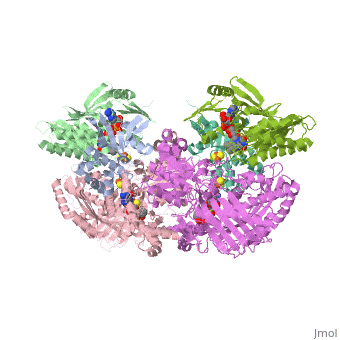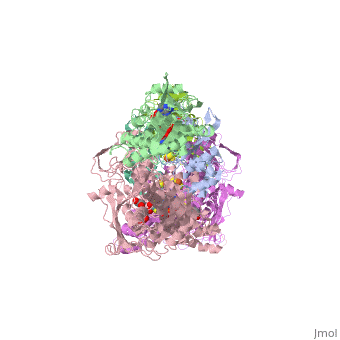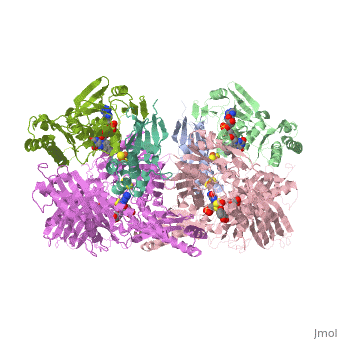
'''CRYSTAL STRUCTURE OF XANTHINE OXIDASE FROM BOVINE MILK'''<br /> | '''CRYSTAL STRUCTURE OF XANTHINE OXIDASE FROM BOVINE MILK'''<br /> | ||
==Overview== | ==Overview== | ||
| - | Mammalian xanthine oxidoreductases, which catalyze the last two steps in | + | Mammalian xanthine oxidoreductases, which catalyze the last two steps in the formation of urate, are synthesized as the dehydrogenase form xanthine dehydrogenase (XDH) but can be readily converted to the oxidase form xanthine oxidase (XO) by oxidation of sulfhydryl residues or by proteolysis. Here, we present the crystal structure of the dimeric (M(r), 290,000) bovine milk XDH at 2.1-A resolution and XO at 2.5-A resolution and describe the major changes that occur on the proteolytic transformation of XDH to the XO form. Each molecule is composed of an N-terminal 20-kDa domain containing two iron sulfur centers, a central 40-kDa flavin adenine dinucleotide domain, and a C-terminal 85-kDa molybdopterin-binding domain with the four redox centers aligned in an almost linear fashion. Cleavage of surface-exposed loops of XDH causes major structural rearrangement of another loop close to the flavin ring (Gln 423Lys 433). This movement partially blocks access of the NAD substrate to the flavin adenine dinucleotide cofactor and changes the electrostatic environment of the active site, reflecting the switch of substrate specificity observed for the two forms of this enzyme. |
==About this Structure== | ==About this Structure== | ||
| - | 1FIQ is a [http://en.wikipedia.org/wiki/Protein_complex Protein complex] structure of sequences from [http://en.wikipedia.org/wiki/Bos_taurus Bos taurus] with FES, MTE, MOS, SAL, FAD and GOL as [http://en.wikipedia.org/wiki/ligands ligands]. Active as [http://en.wikipedia.org/wiki/Xanthine_oxidase Xanthine oxidase], with EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=1.17.3.2 1.17.3.2] Full crystallographic information is available from [http:// | + | 1FIQ is a [http://en.wikipedia.org/wiki/Protein_complex Protein complex] structure of sequences from [http://en.wikipedia.org/wiki/Bos_taurus Bos taurus] with <scene name='pdbligand=FES:'>FES</scene>, <scene name='pdbligand=MTE:'>MTE</scene>, <scene name='pdbligand=MOS:'>MOS</scene>, <scene name='pdbligand=SAL:'>SAL</scene>, <scene name='pdbligand=FAD:'>FAD</scene> and <scene name='pdbligand=GOL:'>GOL</scene> as [http://en.wikipedia.org/wiki/ligands ligands]. Active as [http://en.wikipedia.org/wiki/Xanthine_oxidase Xanthine oxidase], with EC number [http://www.brenda-enzymes.info/php/result_flat.php4?ecno=1.17.3.2 1.17.3.2] Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1FIQ OCA]. |
==Reference== | ==Reference== | ||
| Line 14: | Line 14: | ||
[[Category: Protein complex]] | [[Category: Protein complex]] | ||
[[Category: Xanthine oxidase]] | [[Category: Xanthine oxidase]] | ||
| - | [[Category: Eger, B | + | [[Category: Eger, B T.]] |
[[Category: Enroth, C.]] | [[Category: Enroth, C.]] | ||
[[Category: Nishino, T.]] | [[Category: Nishino, T.]] | ||
[[Category: Okamoto, K.]] | [[Category: Okamoto, K.]] | ||
| - | [[Category: Pai, E | + | [[Category: Pai, E F.]] |
[[Category: FAD]] | [[Category: FAD]] | ||
[[Category: FES]] | [[Category: FES]] | ||
| Line 27: | Line 27: | ||
[[Category: xanthine oxidase]] | [[Category: xanthine oxidase]] | ||
| - | ''Page seeded by [http:// | + | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Thu Feb 21 12:39:06 2008'' |
Revision as of 10:39, 21 February 2008
|
CRYSTAL STRUCTURE OF XANTHINE OXIDASE FROM BOVINE MILK
Overview
Mammalian xanthine oxidoreductases, which catalyze the last two steps in the formation of urate, are synthesized as the dehydrogenase form xanthine dehydrogenase (XDH) but can be readily converted to the oxidase form xanthine oxidase (XO) by oxidation of sulfhydryl residues or by proteolysis. Here, we present the crystal structure of the dimeric (M(r), 290,000) bovine milk XDH at 2.1-A resolution and XO at 2.5-A resolution and describe the major changes that occur on the proteolytic transformation of XDH to the XO form. Each molecule is composed of an N-terminal 20-kDa domain containing two iron sulfur centers, a central 40-kDa flavin adenine dinucleotide domain, and a C-terminal 85-kDa molybdopterin-binding domain with the four redox centers aligned in an almost linear fashion. Cleavage of surface-exposed loops of XDH causes major structural rearrangement of another loop close to the flavin ring (Gln 423Lys 433). This movement partially blocks access of the NAD substrate to the flavin adenine dinucleotide cofactor and changes the electrostatic environment of the active site, reflecting the switch of substrate specificity observed for the two forms of this enzyme.
About this Structure
1FIQ is a Protein complex structure of sequences from Bos taurus with , , , , and as ligands. Active as Xanthine oxidase, with EC number 1.17.3.2 Full crystallographic information is available from OCA.
Reference
Crystal structures of bovine milk xanthine dehydrogenase and xanthine oxidase: structure-based mechanism of conversion., Enroth C, Eger BT, Okamoto K, Nishino T, Nishino T, Pai EF, Proc Natl Acad Sci U S A. 2000 Sep 26;97(20):10723-8. PMID:11005854
Page seeded by OCA on Thu Feb 21 12:39:06 2008
Categories: Bos taurus | Protein complex | Xanthine oxidase | Eger, B T. | Enroth, C. | Nishino, T. | Okamoto, K. | Pai, E F. | FAD | FES | GOL | MOS | MTE | SAL