OhrR
From Proteopedia
| Line 11: | Line 11: | ||
Newbery et al 2007 reported the first crystal structure… ___ to be submitted to PDB… state about redcued mutant | Newbery et al 2007 reported the first crystal structure… ___ to be submitted to PDB… state about redcued mutant | ||
| - | |||
| - | |||
| - | {{STRUCTURE_2pex | PDB=2pex | SCENE= }} | ||
| - | |||
| - | <scene name='User:David_Bruhn/Sandbox1/2pex/1'>TextToBeDisplayed</scene> | ||
| Line 27: | Line 22: | ||
{{STRUCTURE_2pfb | PDB=2pfb | SCENE= }} | {{STRUCTURE_2pfb | PDB=2pfb | SCENE= }} | ||
{{STRUCTURE_2pfb | PDB=2pex | SCENE= }} | {{STRUCTURE_2pfb | PDB=2pex | SCENE= }} | ||
| + | |||
==Transcription Regulation as a Mechanism to Evade Reactive Oxygen Species (ROS)== | ==Transcription Regulation as a Mechanism to Evade Reactive Oxygen Species (ROS)== | ||
Revision as of 04:28, 29 November 2008
|
|
Organic Hydroperoxide Resistance Repressor Protein from Xanthomonas campestris (OhrR Xc)
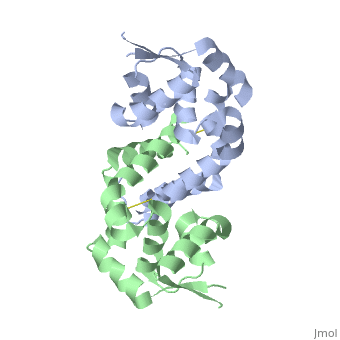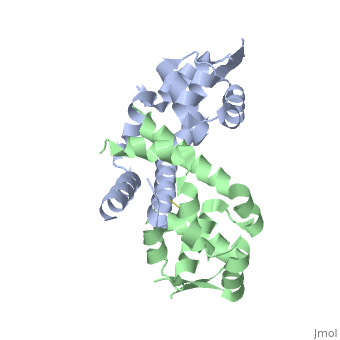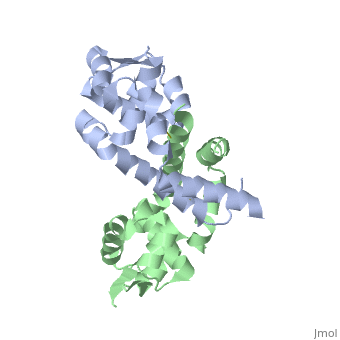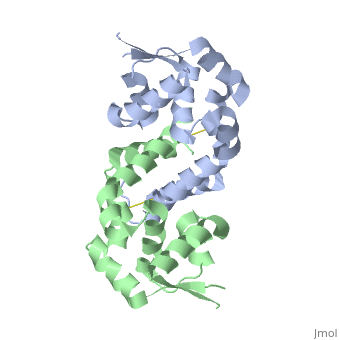
OhrR plays a critical role in sensing of reactive oxygen species in the pathogenic bacteria Xanthomonas campestris. Under normal cellular conditions, dimeric OhrR protein is bound to DNA to repress expression of ohr, which encodes organic hydroperoxide resistance protein. When the reactive oxygen species organic hydroperoxide (OHR) is present in the cell, it oxidizes a reactive cysteine in OhrR, resulting in a conformational change that causes the dissocation of OhrR from ohr and subsequent clearance of OHR by OHR. This dissociation occurs because the oxidation of this reactive cysteine results in the formation of an interprotein cysteine bond between each of two units.
Crystallization of OhrR Xc
Newbery et al 2007 reported the first crystal structure… ___ to be submitted to PDB… state about redcued mutant
Structure of Reduced OhrR Xc
Oxidation-induced Confirmational Changes in OhrR Xc
The image below shows the reduced form of the protein (reds) aligned to the oxidized form (greens). The two most notable structural rearrangements occurring to individual subunits upon activation (oxidation) are labeled. The glycine-induced kink in the middle of α-helix 5 becomes a 3-residue loop region, effectively breaking this helix into 2 (one composed of residues 109-116 and the second composed of 117-129). The formation of a loop in the activated form permits residues 117-129 to rotate and positions C127 to form a disulfide bond with C22’ on α1 of the subunit composing the other half of the dimer. In addition to facilitating inter-subunit disulfide bond formation, the movement of the α-helix 5 also results in a repositioning of α helix 6. Repositioning of α6 facilitates α5 repositioning but does directly stabilize the interaction of the two subunits in the oxidized form.
Template:STRUCTURE 2pfb
Template:STRUCTURE 2pfb
Transcription Regulation as a Mechanism to Evade Reactive Oxygen Species (ROS)
One of several defense methods utilized by the human body is the production of reactive oxygen species in response to the presence of prokaryotic pathogens. Several aspects of the contributions of ROS to the innate immune response have been characterized (Skulachev 1998, Geiszt 2003). The capacity to cope with ROS-driven host cell responses, therefore, represents a mechanism by which pathogens can evade clearance and increase the likelihood of successful pathogenesis. Several pathogens have evolved to exploit such a mechanism, namely through the regulation of ROS-resistance genes by oxidizable transcription factors. The resulting dissociation of the regulator from the target DNA results in expression of gene involved in reduction (and detoxification) of several ROS species.
MarR Family Proteins
One well-studied family of transcription factors involved in regulation of ROS-reducing pathogen genes is the MarR family of proteins. Members of this family of transcriptional regulator have been identified and characterized in several bacterial species including Bacillus subtilis, Staphylococcus aureus, Pseudomonas aeruginosa (SoxR), Escherichia coli (OxyR, Mar) and Xanthomonas campestris. At least one MarR family member (OhrR from Streptomyces coelicolor) is able to serve as a transcriptional regulator when reduced but serve as an activator when oxidized (Oh et al. 2007).