OhrR
From Proteopedia
| Line 13: | Line 13: | ||
==Structure of Reduced OhrR ''Xc''== | ==Structure of Reduced OhrR ''Xc''== | ||
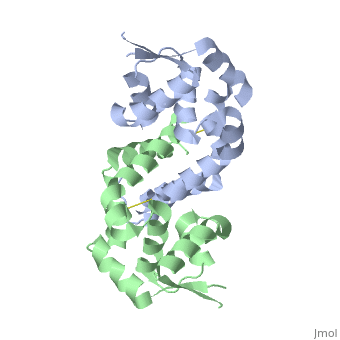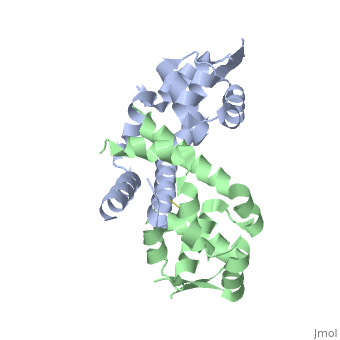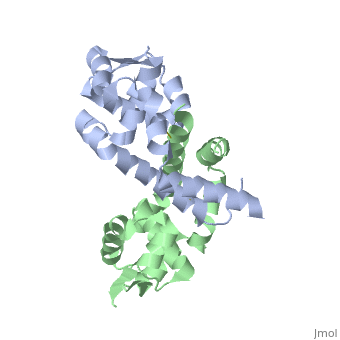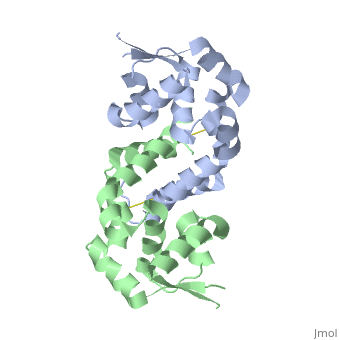
The resulting model 2pex shows the biologically relevant dimer of the protein. Each subunit of the dimer is composed of six α-helices and 3 β-strands. | The resulting model 2pex shows the biologically relevant dimer of the protein. Each subunit of the dimer is composed of six α-helices and 3 β-strands. | ||
| - | The specific residues corresponding to these regions of secondary structure are as follows | + | The specific residues corresponding to these regions of secondary structure are as follows: α1 (residues 21–39), α2 (residues 47–58), β1(residues 62–63), α3 (residues 64–71), α4 (residues 75–87), β2 (residues 91–94), β3 (residues 104–107), α5 (residues 109–129), α6 (residues 133–151), and three-ten helices 1a (residues 13–15) and 1b (residues 17–19). |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
The winged-HTH DNA-binding motif | The winged-HTH DNA-binding motif | ||
- Formed by helices α3 and α4 followed by β strands β2 and β3, which form a small β sheet with β1. | - Formed by helices α3 and α4 followed by β strands β2 and β3, which form a small β sheet with β1. | ||
| - | + | ||
| + | α5 (residues109–129), longest helix | ||
| + | b. displays a kink that is centered about residue G119. | ||
Interface | Interface | ||
Revision as of 01:01, 2 December 2008
|
|
Organic Hydroperoxide Resistance Repressor Protein from Xanthomonas campestris (OhrR Xc)
OhrR plays a critical role in sensing of reactive oxygen species in the pathogenic bacteria Xanthomonas campestris. Under normal cellular conditions, dimeric OhrR protein is bound to DNA to repress expression of ohr, which encodes organic hydroperoxide resistance protein. When the reactive oxygen species organic hydroperoxide (OHR) is present in the cell, it oxidizes a reactive cysteine in OhrR, resulting in a conformational change that causes the dissocation of OhrR from ohr and subsequent clearance of OHR by OHR. This dissociation occurs because the oxidation of this reactive cysteine results in the formation of an interprotein cysteine bond between each of two units.
Crystallization and Quality of OhrR Xc Models
Newbery et al 2007 reported the first crystal structure of OhrR from Xanthomonas campestris in Molecular Cell. This was the second structure of an OhrR protein to be submitted to the Protein Data Bank. For the structures of both reduced and oxidized OhrR, protein was overexpressed in E coli. To produce crystals of the reduced form of the protein, site-directed mutagenesis was performed to mutate the reactive cysteine (Cys22) to a serine. Crystals of both unlabeled and selenomethionine-substituted reduced OhrR were generated and data collected using SAD phasing. For crystallization of the oxidized form of the protein, purified protein was treated with cumene hydroperoxide and purified via gel filtration prior to crystallization. The resulting reduced and oxidized structures were respectively named 2pex and 2pfb. Refinement of 2pex resulted in a 1.90 angstrom structure with an Rfree of 27.7% and Rwork of 23.6% and 96.7% of phi,psi angles in the most favorable regions of the Ramachandran plot. Refinement of 2pfb yielded a 1.93 angstrom structure with 96.1% of phi,psi angles in the most favorable regions of the Ramachandran plot and Rfree/Rwork value of 25.0% and 21.9% respectively. Neither of the final models included any residues in disallowed regions of the Ramachandran plot
Structure of Reduced OhrR Xc
The resulting model 2pex shows the biologically relevant dimer of the protein. Each subunit of the dimer is composed of six α-helices and 3 β-strands. The specific residues corresponding to these regions of secondary structure are as follows: α1 (residues 21–39), α2 (residues 47–58), β1(residues 62–63), α3 (residues 64–71), α4 (residues 75–87), β2 (residues 91–94), β3 (residues 104–107), α5 (residues 109–129), α6 (residues 133–151), and three-ten helices 1a (residues 13–15) and 1b (residues 17–19).
The winged-HTH DNA-binding motif - Formed by helices α3 and α4 followed by β strands β2 and β3, which form a small β sheet with β1.
α5 (residues109–129), longest helix b. displays a kink that is centered about residue G119.
Interface - 310 Helices (1a, 1b) α1, α5, and α6 from each subunit form an extensive dimerization interface that buries a surface area of 5391 Å2
Connecting HTH and OHP sensing domains - Helices α2 and α5 lie perpendicular to each other across one face of each subunit and act to connect the dimerization and OHP sensing domains to the winged-HTH motifs (Figure 1A).
- .
The extensive dimerization domain buries 5391Ų.
Oxidation-induced Confirmational Changes in OhrR Xc
The image below shows the reduced form of the protein (reds) aligned to the oxidized form (greens). The two most notable structural rearrangements occurring to individual subunits upon activation (oxidation) are labeled. The glycine-induced kink in the middle of α-helix 5 becomes a 3-residue loop region, effectively breaking this helix into 2 (one composed of residues 109-116 and the second composed of 117-129). The formation of a loop in the activated form permits residues 117-129 to rotate and positions C127 to form a disulfide bond with C22’ on α1 of the subunit composing the other half of the dimer. In addition to facilitating inter-subunit disulfide bond formation, the movement of the α-helix 5 also results in a repositioning of α helix 6. Repositioning of α6 facilitates α5 repositioning but does directly stabilize the interaction of the two subunits in the oxidized form.
Conserved Residues in OhrR Xc
|
The program Consurf was used to predict which regions of OhrR Xc are most high conserved. The most conserved region of OhrR Xc is the helix-turn-helix region. This is not surprising in that this is a transcription factor and if it is unable to bind DNA, then this function has been severely inhibited. What was surprising, however, is that the reactive cysteine (C22) of the protein and the residues it interacted with (C127) were not highly conserved. The conservation score for C22 was 7 (out of 9) and the C127 was 2 out of 9. Such a low rating for 1 of the 2 available cysteines in the entire protein suggests that disulfide bond formation may not be a conserved mechanism in the homologues included in this query. The regions of the protein that are in contact in the dimer are also conserved, with conservation scores most often between 6 and 7 (out of 10). This is logical in that this interaction interface must be conserved to allow these regions of the protein to facilitate dimerization. The least conserved portions of the protein are those that do not interact with either the DNA opposite unit of the dimer. This is logical in that these regions do not need to conserve a very strict chemistry of geometry in order to serve as a “linker” between the two DNA binding and interaction domain of a individual chain. Residues are colored as.....
Template:STRUCTURE 2pfb Template:STRUCTURE 2pfb
Transcription Regulation as a Mechanism to Evade Reactive Oxygen Species (ROS)
One of several defense methods utilized by the human body is the production of reactive oxygen species in response to the presence of prokaryotic pathogens. Several aspects of the contributions of ROS to the innate immune response have been characterized (Skulachev 1998, Geiszt 2003). The capacity to cope with ROS-driven host cell responses, therefore, represents a mechanism by which pathogens can evade clearance and increase the likelihood of successful pathogenesis. Several pathogens have evolved to exploit such a mechanism, namely through the regulation of ROS-resistance genes by oxidizable transcription factors. The resulting dissociation of the regulator from the target DNA results in expression of gene involved in reduction (and detoxification) of several ROS species.
MarR Family Proteins
One well-studied family of transcription factors involved in regulation of ROS-reducing pathogen genes is the MarR family of proteins. Members of this family of transcriptional regulator have been identified and characterized in several bacterial species including Bacillus subtilis, Staphylococcus aureus, Pseudomonas aeruginosa (SoxR), Escherichia coli (OxyR, Mar) and Xanthomonas campestris. At least one MarR family member (OhrR from Streptomyces coelicolor) is able to serve as a transcriptional regulator when reduced but serve as an activator when oxidized (Oh et al. 2007).