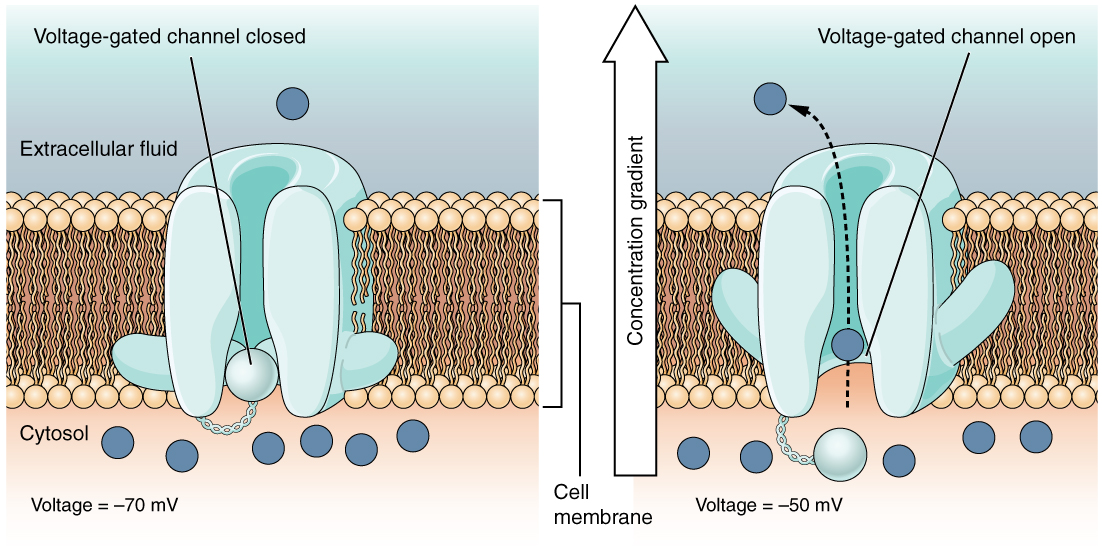
This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Sandbox2qc8
From Proteopedia
(Difference between revisions)
| Line 9: | Line 9: | ||
| - | The beta strands are arranged into five <scene name='Sandbox2qc8/Pdb_defined_beta_strands/1'>beta sheets</scene>. | + | The beta strands are arranged into five <scene name='Sandbox2qc8/Pdb_defined_beta_strands/1'>beta sheets</scene>. In addition, there are 5 <scene name='Sandbox2qc8/Hairpins/1'>beta hairpins</scene> and 5 <scene name='Sandbox2qc8/Bulges/1'>beta bulges</scene>. <ref>European Bioinformatics Institute, Ligase(amide synthetase), http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/pdbsum/GetPage.pl?pdbcode=2gls, Accessed December 18, 2008.</ref> |
| - | . | + | |
The active site within the secondary structure can be called a "bifunnel," providing access to ATP and glutamate at opposing ends.<ref>Eisenberg, D., et al., Structure-function relationships of glutamine synthetases, Biochimica et Biophysica Acta 1477 (2000), 122-145.</ref> | The active site within the secondary structure can be called a "bifunnel," providing access to ATP and glutamate at opposing ends.<ref>Eisenberg, D., et al., Structure-function relationships of glutamine synthetases, Biochimica et Biophysica Acta 1477 (2000), 122-145.</ref> | ||
| Line 16: | Line 15: | ||
The only ligand present is a pair of Mn ions (Manganese) that indicates the active site of each subunit of the dodecamer. | The only ligand present is a pair of Mn ions (Manganese) that indicates the active site of each subunit of the dodecamer. | ||
| - | <scene name='Sandbox2qc8/Hairpins/1'>Hairpins</scene>, | ||
| - | <scene name='Sandbox2qc8/Bulges/1'>Bulges</scene>, | ||
<scene name='Sandbox2qc8/Catalytic_sites/1'>Catalytic sites E327, R339, D50</scene><br> | <scene name='Sandbox2qc8/Catalytic_sites/1'>Catalytic sites E327, R339, D50</scene><br> | ||
Revision as of 03:15, 19 December 2008
Glutamine Synthetase: Secondary structures
Glutamine synthetase is composed of 12 . Each subunit is composed of 15 and . Each subunit binds 2 Mn for a total of per Glutamine Synthetase.
Each subunit has an exposed NH2 terminus and buried COOH terminus as part of a . [1]
The beta strands are arranged into five . In addition, there are 5 and 5 . [2]
The active site within the secondary structure can be called a "bifunnel," providing access to ATP and glutamate at opposing ends.[3]
The only ligand present is a pair of Mn ions (Manganese) that indicates the active site of each subunit of the dodecamer.
References
- ↑ Yamashita, M., et al.,Refined Atomic Model of Glutamine Synthetase at 3.5A Resolution, The Journal of Biological Chemistry, 1989, 17681-17690.
- ↑ European Bioinformatics Institute, Ligase(amide synthetase), http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/pdbsum/GetPage.pl?pdbcode=2gls, Accessed December 18, 2008.
- ↑ Eisenberg, D., et al., Structure-function relationships of glutamine synthetases, Biochimica et Biophysica Acta 1477 (2000), 122-145.