User:Lois A. Fridmann/Sandbox 3
From Proteopedia
| Line 22: | Line 22: | ||
==Mutations - PTAP binding pocket, ubiquitin binding pocket, C-terminal UEV Domain== | ==Mutations - PTAP binding pocket, ubiquitin binding pocket, C-terminal UEV Domain== | ||
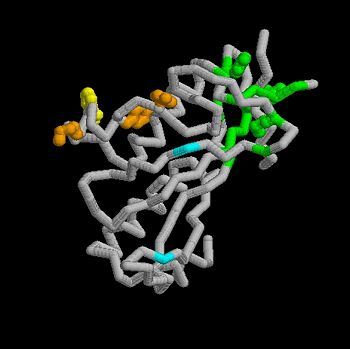
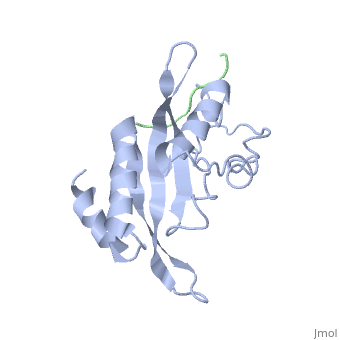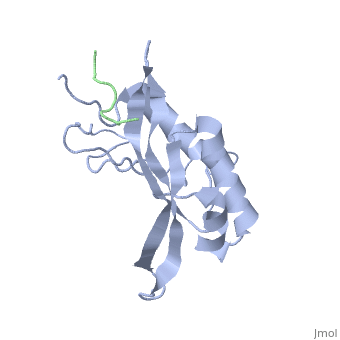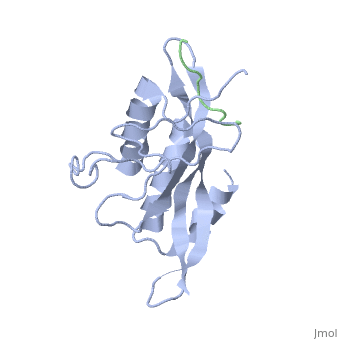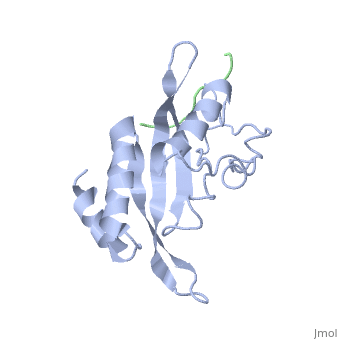
| + | The Tsg101 UEV domain with specific highlights of the PTAP binding site featured as a green backbone and three residues in the PTAP binding site (<scene name='User:Lois_A._Fridmann/Sandbox_3/M95/2'>M95</scene>, <scene name='User:Lois_A._Fridmann/Sandbox_3/Y63/1'>Y63</scene>, <scene name='User:Lois_A._Fridmann/Sandbox_3/Y68/1'>Y68</scene>), which are also colored green. Four C-terminal domain residues <scene name='User:Lois_A._Fridmann/Sandbox_3/Y110/1'>Y110</scene>, <scene name='User:Lois_A._Fridmann/Sandbox_3/Y113/1'>Y113</scene>, <scene name='User:Lois_A._Fridmann/Sandbox_3/K118/1'>K118</scene> colored orange and and <scene name='User:Lois_A._Fridmann/Sandbox_3/W117/1'>W117</scene> colored yellow, and two residues included in the Ubiquitin binding site featured as a cyan colored backbone (<scene name='User:Lois_A._Fridmann/Sandbox_3/Y43/1'>V43</scene>, <scene name='User:Lois_A._Fridmann/Sandbox_3/F88/1'>F88</scene>). | ||
| - | The | + | The three residues featured in the PTAP binding pocket exhibit potential areas where mutations can occur that results in the elimination or reduction of the HIV-1 Gag protein binding. The two residues featured in the Ubiquitin binding pocket feature potential areas where mutations can occur that inhibits Ubiquitination of the Tsg101 or the p6 region of the Gag binding to the PTAP binding pocket. |
| - | The Tsg101 UEV domain with specific highlights of the PTAP binding site featured as a green backbone and three residues in the PTAP binding site (M95, Y63, Y68), which are also colored green. Four C-terminal domain residues Y110, Y113, K118 colored orange and W117 colored yellow, and two residues included in the Ubiquitin binding site featured as a cyan colored backbone (V43, F88). | ||
| - | The three residues featured in the PTAP binding pocket exhibit potential areas where mutations can occur that results in the elimination or reduction of the HIV-1 Gag protein binding. The two residues featured in the Ubiquitin binding pocket feature potential areas where mutations can occur that inhibits Ubiquitination of the Tsg101 or the p6 region of the Gag binding to the PTAP binding pocket. | ||
However, the main focus of the model is the C-terminal region of the UEV domain where the mutation of certain amino acid residues, which are not directly involved in the PTAP binding pocket result in the elimination or reduction of binding to the Gag protein. A mutation of W117 (yellow residues), which is conserved in all E2 enzymes, to A117 eliminates binding entirely. Mutations of Y110, Y113 and K118 (orange residues), which are unique to the Tsg101, to W110, V113 and A118 reduce binding by 31%, 33% and 50% respectively. | However, the main focus of the model is the C-terminal region of the UEV domain where the mutation of certain amino acid residues, which are not directly involved in the PTAP binding pocket result in the elimination or reduction of binding to the Gag protein. A mutation of W117 (yellow residues), which is conserved in all E2 enzymes, to A117 eliminates binding entirely. Mutations of Y110, Y113 and K118 (orange residues), which are unique to the Tsg101, to W110, V113 and A118 reduce binding by 31%, 33% and 50% respectively. | ||
| - | |||
Identification of mutations of various residues that inhibits HIV-1 Gag binding can provide important information and avenues for future research aimed at discovery of new anti-viral agents. This model represents the wild type version of Tsg101 UEV domain and it highlights residues that contact P(S/T)AP and/or residues found by mutational analysis, which are important in the interaction. The residues fall into four groups: residues in the P(S/T)AP binding pocket - M95, Y63, Y68 (green); residues in the Ub binding pocket (2 critical residues are highlighted in the model) - V43, F88 (cyan); residues unique to Tsg101 that are not conserved in related E2 enzymes - Y110, Y113, K118 (orange); and residues conserved in Tsg101 and related E2 enzymes - W117 (yellow). | Identification of mutations of various residues that inhibits HIV-1 Gag binding can provide important information and avenues for future research aimed at discovery of new anti-viral agents. This model represents the wild type version of Tsg101 UEV domain and it highlights residues that contact P(S/T)AP and/or residues found by mutational analysis, which are important in the interaction. The residues fall into four groups: residues in the P(S/T)AP binding pocket - M95, Y63, Y68 (green); residues in the Ub binding pocket (2 critical residues are highlighted in the model) - V43, F88 (cyan); residues unique to Tsg101 that are not conserved in related E2 enzymes - Y110, Y113, K118 (orange); and residues conserved in Tsg101 and related E2 enzymes - W117 (yellow). | ||
Revision as of 01:13, 8 June 2009
HIV-1 Gag Recruitment of Tsg101 and the Viral Budding Process
Contents |
Background and History
The Human Immunodefiency Virus (HIV) is the causative agent of acquired immunodefiency syndrome (AIDS) in which the immune system of a human begins to fail, leading to life threatening infections. HIV is a retrovirus that uses its host’s cellular machinery to replicate. The Tumor susceptibility gene 101 [Tsg101], is a human gene that encodes for a cellular protein, plays an important role in the pathogenesis of HIV and belongs to the ubiquitin-conjugating enzyme family, the ubiquitin E2 variant (UEV) subfamily.
In normal functioning cells, Tsg101, as a subunit of the ESCRT-1 (Endosomal Sorting Complex Required for Transport), promotes membrane alterations inside the cell that result in the formation of compartments (multivesicular bodies) in which proteins that have been marked for transport to the lysosome, through ubiquination, are sorted. Normally, a specific tetrapeptide PSAP binding motif on the endosomal protein Hrs (hepatocyte growth factor-regulated tyrosine kinase substrate) will bind to the specific P(S/T)AP binding pocket on the Tsg101 and deliver the protein cargo to endosomes that eventually become or fuse with lysosomes. If HIV is present in a cell, a major structural protein of the virus, Gag, recruits the Tsg101 protein using its own PTAP motif, which mimics the Hrs and binds to the binding pocket of the Tsg101 to gain access to the downstream machinery and to mediate viral budding as an alternative to degradation. The Gag protein is a Group-specific antigen and contains the genetic material that codes for core structural proteins of retroviruses. PTAP and PSAP motifs are able to bind Tsg101 equally.
Poly-ubiquitination inactivates Tsg101, possibly by altering the shuttling of a active membrane-bound protein with an inactive soluble protein.
| |||||||||
| 1m4p, 20 NMR models () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene: | tumor susceptibility gene 101 (Homo sapiens), Gag (Human immunodeficiency virus 1) | ||||||||
| Related: | 1kpp, 1kpq, 1m4q | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Structural and Functional Properties of the Tsg101
The features the wild type version of the , amino acid residues Met 1 through Pro 145 and the protein, amino acid residues Pro 205 through Glu 213. Focusing on the Tsg101 UEV domain of this PDB, important features of this protein are observed that may be directly involved in the normal trafficking or viral budding processes. The Tsg101 is referred to as a UEV domain due to it’s overall α/β/α/ loop configuration that is similar to other E2 ubiquitin ligases, but it does not have the C-terminal helices found in many other E2 enzymes. The Tsg101 C-terminal end is referred to as the C-terminal end of the UEV domain because the precise C-terminal end of the Tsg101 protein has not been determined at this time.
The Tsg101 UEV domain features the represented by amino acid residues: Thr 58; Val 61; Tyr 63; Tyr 68; Asn 69; Ile 70; Pro 71; Thr 92; Met 95; Pro 139; Val 141; Phe 142; Ser 143 and Arg 144. This binding pocket, which normally binds the protein Hrs but can also bind the HIV-1 Gag protein equally.
Another important feature on the Tsg101 that may be involved in the normal trafficking or viral budding process is the ubiquitin-binding pocket. The of the Tsg101 UEV domain has eight specific residues that are involved: residues 43 through 49; and residue 88. The flanking or end domains are Valine 43 and Phenylalanine 88. These residues are highlighted because they mark the ends of the ubiquitin-binding pocket, where ubiquitin binds to help escort the HIV-1 Gag into the trafficking lane and onto the membrane for budding. When the Gag is bound to the PTAP the ubiquitin-binding pocket does not appear to bind more than one ubiquitin. If more than four are bound the Tsg101 is then marked for degradation.
A third structural feature of the Tsg101 UEV domain that may be indirectly involved in the overall trafficking and viral budding is amino acid residues occupying the C-terminal end of the UEV domain. Mutations in this area are discussed below.
Mutations - PTAP binding pocket, ubiquitin binding pocket, C-terminal UEV Domain
The Tsg101 UEV domain with specific highlights of the PTAP binding site featured as a green backbone and three residues in the PTAP binding site (, , ), which are also colored green. Four C-terminal domain residues , , colored orange and and colored yellow, and two residues included in the Ubiquitin binding site featured as a cyan colored backbone (, ).
The three residues featured in the PTAP binding pocket exhibit potential areas where mutations can occur that results in the elimination or reduction of the HIV-1 Gag protein binding. The two residues featured in the Ubiquitin binding pocket feature potential areas where mutations can occur that inhibits Ubiquitination of the Tsg101 or the p6 region of the Gag binding to the PTAP binding pocket.
However, the main focus of the model is the C-terminal region of the UEV domain where the mutation of certain amino acid residues, which are not directly involved in the PTAP binding pocket result in the elimination or reduction of binding to the Gag protein. A mutation of W117 (yellow residues), which is conserved in all E2 enzymes, to A117 eliminates binding entirely. Mutations of Y110, Y113 and K118 (orange residues), which are unique to the Tsg101, to W110, V113 and A118 reduce binding by 31%, 33% and 50% respectively.
Identification of mutations of various residues that inhibits HIV-1 Gag binding can provide important information and avenues for future research aimed at discovery of new anti-viral agents. This model represents the wild type version of Tsg101 UEV domain and it highlights residues that contact P(S/T)AP and/or residues found by mutational analysis, which are important in the interaction. The residues fall into four groups: residues in the P(S/T)AP binding pocket - M95, Y63, Y68 (green); residues in the Ub binding pocket (2 critical residues are highlighted in the model) - V43, F88 (cyan); residues unique to Tsg101 that are not conserved in related E2 enzymes - Y110, Y113, K118 (orange); and residues conserved in Tsg101 and related E2 enzymes - W117 (yellow). Three residues in the P(S/T)AP binding pocket are sites where mutations eliminate or reduce P(S/T)AP binding. The two residues in the Ub binding pocket are sites where mutations inhibit binding of a Ub-modified peptide derived from the PTAP-containing region of Gag. The model also highlighted a region at the C-terminal end of the UEV domain where mutations of certain amino acids residues that are not directly involved in the P(S/T)AP binding pocket resulted in the elimination or reduction of binding. Mutation of W117, which is conserved in all E2 enzymes, eliminated binding completely. Mutation of Y110, Y113 and K118, which are unique to Tsg101, reduced binding to 31%, 33% and 50% respectively, of the binding observed for the non-mutated UEV domain.
Future Research
There are a number of sites on Tsg101 that are involved in normal trafficking or viral budding functions (VerPlank et al. 2001, Pornillos et al. 2002). The binding of HIV-1 Gag to the PTAP binding pocket works in coordination with the binding of the ubiquitin. Perhaps, when HIV-1 Gag is bound in the pocket, Gag containing more than one ubiquitin molecule cannot bind in the P(S/T)AP binding pocket or more than one Ub molecule cannot be added to Gag, thereby preventing Gag from being sorted to the normal degradation pathway.
BIAcore biosensor experiments conducted by Pornillos, et al. 2003, showed that the binding affinities of the P(S/T)AP motifs in wild type and mutated HIV-1 Gag p6 and Hrs exhibit variation. Binding affinity was greatest for the sequence in HIV-1 Gag as compared with Hrs and the mutated HIV-1 Gag. The binding affinities were shown to be directly attributed to Tsg101-Gag binding because when the proline was mutated to leucine, no binding was detected. Specific sequences flanking the central P(S/T)AP motif appeared to affect the absolute binding affinities between Tsg101 and Hrs. These data suggest that when specific residues located in the PTAP binding pocket of Tsg101 are mutated, there will be a reduction in the binding affinity not only for the HIV-1 Gag, but also for other cellular proteins such as Hrs. In addition to these binding affinity data a specific 8-member cyclic peptide, which was selected for in an experiment conducted by Tavassoli, et al. 2008, was shown to decrease the interaction of both Gag-Tsg101 and Hrs-Tsg101. This further supports the importance of understanding the interaction of Gag-Tsg101 in the P(S/T)AP binding site and flanking residues sites, and exhibits the possibility of inhibiting specific protein-protein interactions which may lead to viral budding.
Tsg101 UEV domain residues Y110, Y113, and K118 are unique to Tsg101, while W117 is conserved in E2 enzymes and Tsg101. Mutations in all of these specific sites appear to cause a significant decrease in Gag interaction with the P(S/T)AP binding pocket. In the case of the unique residues, the alteration may have altered sites critical for the structure of the catalytically unresponsive active site in Tsg101, which as noted, is a variant form of the E2 enzyme. In the case of the conserved W117 residue, mutations may have disrupted a site important for overall protein folding or stability. Most significant is the fact that these residues affect binding, which suggests that anti-viral strategies currently being developed against SH3 and WW domain protein-binding motifs might be employed. As in Tsg101, critical binding determinants are regions containing aromatic residues (in Tsg101 these are Y110, Y113 and W117), which are flanked by charged residues (in Tsg101 these are K108 and K118). This decrease may be attributed to significant conformational changes within the Tsg101 protein that occurs when different steric effects are experienced either through mutations or other binding peptides. Polymers of N-substituted glycine (NSG) residues have recently emerged as an important class of peptide inhibitors that effectively can substitute for proline in WW and SH3 domain-binding proteins. The similarities between Tsg101 and SH3 domains in their recognition of key proline residues suggest that the replacement of proline residues with NSG constructs may eventually lead to the preparation of PTAP-based Tsg101 binding inhibitors that can potentially prevent viral budding. Our physical model illustrates clearly the relationship between these two important regions of the Tsg101 UEV domain (colored orange and yellow) and the other two critical binding regions (colored green and cyan).
References
1. Liu F, Stephen Ag, Adamson C, Gousset K, Aman MJ, Freed EO, Fisher RJ, Burke TR, Jr. 2005. Hydrazone and Hydrazide-Containing N-Substituted Glycines as Peptoid Surrogates for Expedited Library Synthesis: Application to the Preparation of Tsg101-Directed HIV-1 Budding Antagonist. Org Lett 8: 5163-5168.
2. Macias M, Wiesnera S, Sudolb M. 2002. WW and SH3 domains, two different scaffolds to recognize proline-rich ligands. Protein Domains 513:30-37.
3. Pornillos O, Alam S, Davis D, Sundquist W. 2002. Structure of the Tsg101 UEV domain in complex with the PTAP motif of the HIV-1 p6 protein. Nature Structural Biology 9:812 – 817.
4. Pornillos O, Higginson D, Stray K, Fisher R, Garrus J, Payne M, He G, Wang H, Morham S, Sundquist W. 2003. HIV Gag mimics the Tsg101-recruiting activity of the human Hrs protein. The Journal of Cell Biology 162: 425-434.
5. Tavassoli A, Lu Q, Gam J, Pan H, Benkovic S, Cohen S. 2008. Inhibition of HIV Budding by a Genetically Selected Cyclic Peptide Targeting the Gag-TSG101 Interaction. American Chemical Society, Chemical Biology 3:757 - 764.
6. VerPlank L, Bouamr F, LaGrassa T, Agresta B, Kikonyogo A, Leis J, Carter C. 2001. Tsg101, a homolgue of ubiquitin-conjugating (E2) enzymes, binds the L domain of HIV type 1 Pr55Gag. Proceeding of the National Academy of Science 98:7724 – 7729.