User:Ramiro Barrantes/FpgNeiRepair
From Proteopedia
(→Site-directed mutants) |
(→Site-directed mutants) |
||
| Line 148: | Line 148: | ||
| <scene name='User:Ramiro_Barrantes/Workbench/Econeiarginine171/3' target="ecoNeiMutants">EcoNei R171</scene> || ''E. coli'' Nei ([[1k3w]])|| reduced activity, loss of opposite base discrimination || <ref>PMID:17002303</ref> | | <scene name='User:Ramiro_Barrantes/Workbench/Econeiarginine171/3' target="ecoNeiMutants">EcoNei R171</scene> || ''E. coli'' Nei ([[1k3w]])|| reduced activity, loss of opposite base discrimination || <ref>PMID:17002303</ref> | ||
|- | |- | ||
| - | | <scene name='User:Ramiro_Barrantes/Workbench/Econeid174/1' target="ecoNeiMutants">EcoNei | + | | <scene name='User:Ramiro_Barrantes/Workbench/Econeid174/1' target="ecoNeiMutants">EcoNei E174Q</scene> || ''E. coli'' Nei ([[1k3w]])|| ok lyase, reduced glycosylase || <ref>PMID:11711552</ref> |
|- | |- | ||
| <scene name='User:Ramiro_Barrantes/Workbench/Econeir212/1' target="ecoNeiMutants">EcoNei R212A</scene> || ''E. coli'' Nei ([[1k3w]]) || Decreased activity on 5S,6R Tg, slightly less active on DHU|| <ref>PMID:11847126</ref> | | <scene name='User:Ramiro_Barrantes/Workbench/Econeir212/1' target="ecoNeiMutants">EcoNei R212A</scene> || ''E. coli'' Nei ([[1k3w]]) || Decreased activity on 5S,6R Tg, slightly less active on DHU|| <ref>PMID:11847126</ref> | ||
Revision as of 21:59, 16 June 2009
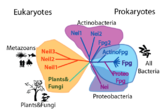
The FpgNei Protein Superfamily
Background on DNA Repair
The genome of any living organisms is being continuously affected by exogenous and endogenous agents, such as ultraviolet light, ionizing radiation, different chemicals and the cell's own metabolites (such as reactive oxygen). Therefore, different systems have evolved to repair these damages, with some of these systems shared throughout all lifeforms. Therefore, the proper functioning of DNA repair is critical for survival. There are six pathways of DNA repair (reviewed in Friedberg et al), and one of them is base-excision repair. The latter's distinguishing feature is that it removes lesions as single bases, as opposed to dNMPs or short oligonucleotides like other systems. [1]
Base excision repair's signature enzyme are the DNA glycosylases. These enzymes work by recognizing and removing a single damaged base from DNA. They are called DNA glycosylases because they hydrolize the N-glycosidic bond of the damaged deoxynucleoside. The subsequent steps of the pathway (strand incision, gap-filling and ligation) are done by other enzymes. [2][3]
Background on Fpg Nei
Overall Function and Structure
| (For FPG, the structure used was 1r2y and for Nei 1k3w) Members of this family have . DNA binds in a cleft between the domains. If EcoNei (structure 1k3w, , and (Thymine Glycol in the case of Nei). [4]. Some of these amino acids are stabilized by a (although a zincless finger motif is present in some of these subfamilies [5]). Other key components are the intercalation loop (different between and ), a group of amino acids that insert into the vacated spot left by the "flipped" base; and amino acids which contact the opposite strand and confer opposite-base specificity. Note that the last two elements discussed, the zinc vs. zincless finger, and the two kinds of intercalation loops, are examples of coevolving functional clusters, groups of amino acids that perform a function, and that might be unnecessary or compensated for within the other subfamilies. We developed a novel method for identifying these clusters and have applied it to bring insight into the structure, function and evolution FpgNei family.
Several authors have suggested mechanisms for these enzymes, please see references for more information [6][7][8][9]. Homologous structures have been solved, including Fpg protein from Lactococcus Lactis (1pjj)[10], Bacillus Stereothermophilus (1r2y)[11], Thermos Thermophilus (1ee8)[12] and Escherichia Coli(1k82)[13] and Nei from Escherichia Coli (1k3w)[14]. The overall structure is similar, and some of the damages include 8-oxoguanine and fapyG (1xc8)[15]. |
|
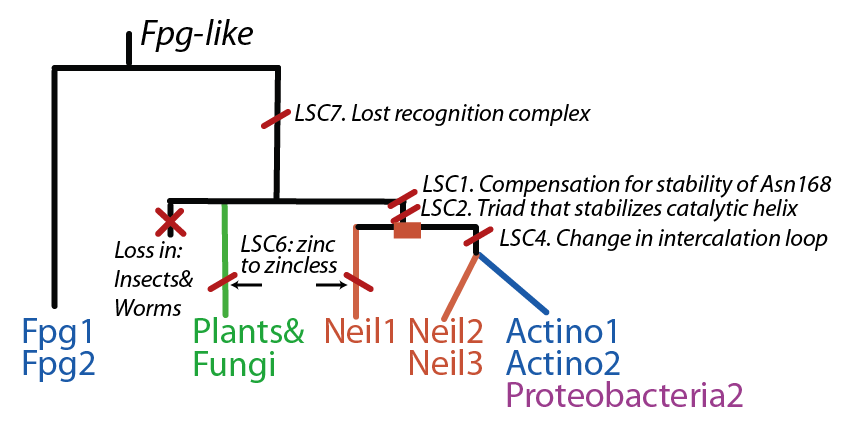
Latent Structural Characters (LSCs)
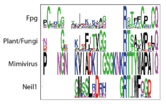
We define a functional cluster as neighboring amino acids which have changed in rate or constraint for a given clade or set of clades. For example, the cysteines in the zinc finger are all conserved in the fpg1, fpg2, actinobacterial and eukaryotic clades, but have a higher rate in plants and neil1, as the latter have a zinc-less finger (see here for more information). We developed a novel method to find these groups of amino acids, discovering previously unknown groups of amino acids which be functionally important in some of these subfamilies.
The table below has some of our main findings, there are two variants for each functional cluster in case that for one functional cluster there is a compensating cluster in the other subfamilies. If you click on one of them you will see the result in the structure, and you can go to the explanation and the sequence distribution below (we have found it helpful to open a separate screen to go along the structure).
|
|
| Functional Cluster | Variant 1 | Variant 2 | Fpg1 | Fpg2 | Plant | Neil1 | Neil2 | Neil3 | Proteo | Actino1 | Actino2 | MimiVirus |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Support for perfectly conserved Asn174 | Y | Y | N | N | N | N | N | N | N | N | ||
| Stability of catalytic helix | Y | Y | Y | N | N | N | N | N | N | N | ||
| Stability of intercalation loop | Y | Y | Y | N | N | N | N | N | N | Y | ||
| Intercalation loop | Y | Y | Y | N | N | N | N | N | N | Y | ||
| Zinc Finger | Y | Y | N | N | Y | Y | Y | Y | Y | N | ||
| Recognition complex | none | Y | N | N | N | N | N | N | N | N | N | |
| Neil1-specific: Support for Lys60 | Y | Y | Y | N | Y | Y | Y | Y | Y | Y | ||
| PlantFungi-specific: R254 DNA binding | different in plants | Y | Y | N | Y | Y | Y | N | Y | Y | Y |
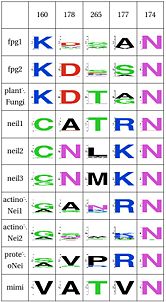
LSC1: Stability of perfectly conserved Asn168 ( and ). Asn174, along with two other amino acids have an effect in the orientation and kinking of the DNA [16], keeping the proper arrangement between the zinc finger and the H2TH [17]. In 4 of the 9 clades (Fpg1, Fpg2 and Plants and Fungi) Asn174 is supported by the amino acid corresponding to Lys160, which in turn hydrogen bonds with Leu249 and Ser250. In the other clades (Actinobacteria 1 and 2, Proteacteria and all vertebrate subfamilies), this role is played by Arg171, which originates on a different helix. The Zinc Finger is shaped differently in the absence of DNA, and there is a hydrogen bond between one of the beta-sheets and the arginine. Site directed mutagenesis has been performed on both Lys155 (corresponding lysine in E. coli Fpg) and as well as Arg171, with the first one being associated with premature dissociation and loss of activity and the latter with significant loss of activity [18][19].
|
LSC2: Stability of catalytic helix( and ) The triad Leu4, Glu8 and Arg57 interact and provide stability to helixA, which has the catalytic residue Pro2,Glu3 and Glu6 [20][21]. This triad is present in the same four clades as above (Fpg1, Fpg2 and Plants and Fungi) but absent in the remaining clades and it is not clear how the same stability is provided. Leu211 also has a hydrophobic interaction with Leu4. The role of Gly4 is not known
|
LSC3: Stability of intercalation loopImage:IntercalationLoopSupport.jpg This structure provides stability for the amino acids that insert into the space vacated by the damaged base |
LSC4: Stability of key Gly59 and Lys60Gly59 and Lys60 are important in the activity of MutM. We hypothesize that Glu137 is very important to maintain its stability, this amino acid is compensated by Asn172 in Neil1 </ref> |
LSC5: Intercalation LoopImage:IntercalationLoop.jpg The residue in positions 77 and 78 suggest a possible intercalation loop The residue E2 and E6 have been mutated, with the first one inactivating the protein and the second one having no major effect [22]. |
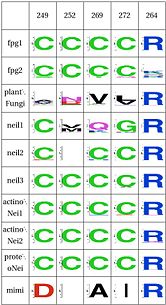
LSC6: Zinc/zincless fingerThe Zinc finger (of the β/β-antiparallel CCCC type) serves to hold the absolutely conserved Arg264 residue [23], which binds to the phosphate of the damaged base and is crucial for function (mutation of this site results in failed cleavage of the damaged base [24]). Site directed mutagenesis experiments have been performed on all four cysteines, leading to loss of activity [25][26], emphasizing the importance of this LSC. In Neil1, there is no Zinc but there is an equivalent structure: a "zincless finger" [27]. Both the plants and mimivirus have a zincless finger as well [28][29], although it is not clear if these are all homologous. |
LSC7: Recognition ComplexThis complex is key in recognizing a damaged guanine[30] |
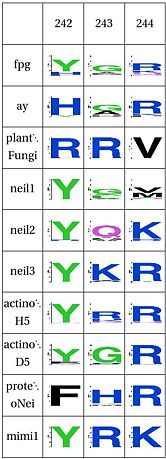
LSC8: DNA binding Tyrosine |
Zinc/zincless finger
Evolution
The FpgNei evolution has not been easy to resolve [31], especially in the deeper branches. Assuming that functional clusters evolve more slowly than individual residues, we can use this as phylogenetic characters to 1) draw the most parsimonious evolution of the superfamily as dictated by these functional clusters 2) examine how these clusters have evolved and how this might have influenced the evolution of FpgNei.
Site-directed mutants
|
|
References
- ↑ Zharkov DO. Base excision DNA repair. Cell Mol Life Sci. 2008 May;65(10):1544-65. PMID:18259689 doi:10.1007/s00018-008-7543-2
- ↑ Zharkov DO. Base excision DNA repair. Cell Mol Life Sci. 2008 May;65(10):1544-65. PMID:18259689 doi:10.1007/s00018-008-7543-2
- ↑ Robertson AB, Klungland A, Rognes T, Leiros I. DNA repair in mammalian cells: Base excision repair: the long and short of it. Cell Mol Life Sci. 2009 Mar;66(6):981-93. PMID:19153658 doi:10.1007/s00018-009-8736-z
- ↑ Zharkov DO, Golan G, Gilboa R, Fernandes AS, Gerchman SE, Kycia JH, Rieger RA, Grollman AP, Shoham G. Structural analysis of an Escherichia coli endonuclease VIII covalent reaction intermediate. EMBO J. 2002 Feb 15;21(4):789-800. PMID:11847126 doi:10.1093/emboj/21.4.789
- ↑ Doublie S, Bandaru V, Bond JP, Wallace SS. The crystal structure of human endonuclease VIII-like 1 (NEIL1) reveals a zincless finger motif required for glycosylase activity. Proc Natl Acad Sci U S A. 2004 Jul 13;101(28):10284-9. Epub 2004 Jul 1. PMID:15232006 doi:10.1073/pnas.0402051101
- ↑ Zharkov DO, Golan G, Gilboa R, Fernandes AS, Gerchman SE, Kycia JH, Rieger RA, Grollman AP, Shoham G. Structural analysis of an Escherichia coli endonuclease VIII covalent reaction intermediate. EMBO J. 2002 Feb 15;21(4):789-800. PMID:11847126 doi:10.1093/emboj/21.4.789
- ↑ Sugahara M, Mikawa T, Kumasaka T, Yamamoto M, Kato R, Fukuyama K, Inoue Y, Kuramitsu S. Crystal structure of a repair enzyme of oxidatively damaged DNA, MutM (Fpg), from an extreme thermophile, Thermus thermophilus HB8. EMBO J. 2000 Aug 1;19(15):3857-69. PMID:10921868 doi:http://dx.doi.org/10.1093/emboj/19.15.3857
- ↑ Pereira de Jesus K, Serre L, Zelwer C, Castaing B. Structural insights into abasic site for Fpg specific binding and catalysis: comparative high-resolution crystallographic studies of Fpg bound to various models of abasic site analogues-containing DNA. Nucleic Acids Res. 2005 Oct 20;33(18):5936-44. Print 2005. PMID:16243784 doi:http://dx.doi.org/33/18/5936
- ↑ Gilboa R, Zharkov DO, Golan G, Fernandes AS, Gerchman SE, Matz E, Kycia JH, Grollman AP, Shoham G. Structure of formamidopyrimidine-DNA glycosylase covalently complexed to DNA. J Biol Chem. 2002 May 31;277(22):19811-6. Epub 2002 Mar 23. PMID:11912217 doi:http://dx.doi.org/10.1074/jbc.M202058200
- ↑ Pereira de Jesus K, Serre L, Zelwer C, Castaing B. Structural insights into abasic site for Fpg specific binding and catalysis: comparative high-resolution crystallographic studies of Fpg bound to various models of abasic site analogues-containing DNA. Nucleic Acids Res. 2005 Oct 20;33(18):5936-44. Print 2005. PMID:16243784 doi:http://dx.doi.org/33/18/5936
- ↑ Fromme JC, Verdine GL. DNA lesion recognition by the bacterial repair enzyme MutM. J Biol Chem. 2003 Dec 19;278(51):51543-8. Epub 2003 Oct 1. PMID:14525999 doi:10.1074/jbc.M307768200
- ↑ Sugahara M, Mikawa T, Kumasaka T, Yamamoto M, Kato R, Fukuyama K, Inoue Y, Kuramitsu S. Crystal structure of a repair enzyme of oxidatively damaged DNA, MutM (Fpg), from an extreme thermophile, Thermus thermophilus HB8. EMBO J. 2000 Aug 1;19(15):3857-69. PMID:10921868 doi:http://dx.doi.org/10.1093/emboj/19.15.3857
- ↑ Gilboa R, Zharkov DO, Golan G, Fernandes AS, Gerchman SE, Matz E, Kycia JH, Grollman AP, Shoham G. Structure of formamidopyrimidine-DNA glycosylase covalently complexed to DNA. J Biol Chem. 2002 May 31;277(22):19811-6. Epub 2002 Mar 23. PMID:11912217 doi:http://dx.doi.org/10.1074/jbc.M202058200
- ↑ Zharkov DO, Golan G, Gilboa R, Fernandes AS, Gerchman SE, Kycia JH, Rieger RA, Grollman AP, Shoham G. Structural analysis of an Escherichia coli endonuclease VIII covalent reaction intermediate. EMBO J. 2002 Feb 15;21(4):789-800. PMID:11847126 doi:10.1093/emboj/21.4.789
- ↑ Coste F, Ober M, Carell T, Boiteux S, Zelwer C, Castaing B. Structural basis for the recognition of the FapydG lesion (2,6-diamino-4-hydroxy-5-formamidopyrimidine) by formamidopyrimidine-DNA glycosylase. J Biol Chem. 2004 Oct 15;279(42):44074-83. Epub 2004 Jul 10. PMID:15249553 doi:10.1074/jbc.M405928200
- ↑ Zharkov DO, Golan G, Gilboa R, Fernandes AS, Gerchman SE, Kycia JH, Rieger RA, Grollman AP, Shoham G. Structural analysis of an Escherichia coli endonuclease VIII covalent reaction intermediate. EMBO J. 2002 Feb 15;21(4):789-800. PMID:11847126 doi:10.1093/emboj/21.4.789
- ↑ Sugahara M, Mikawa T, Kumasaka T, Yamamoto M, Kato R, Fukuyama K, Inoue Y, Kuramitsu S. Crystal structure of a repair enzyme of oxidatively damaged DNA, MutM (Fpg), from an extreme thermophile, Thermus thermophilus HB8. EMBO J. 2000 Aug 1;19(15):3857-69. PMID:10921868 doi:http://dx.doi.org/10.1093/emboj/19.15.3857
- ↑ Rabow L, Venkataraman R, Kow YW. Mechanism of action of Escherichia coli formamidopyrimidine N-glycosylase: role of K155 in substrate binding and product release. Prog Nucleic Acid Res Mol Biol. 2001;68:223-34. PMID:11554299
- ↑ Kropachev KY, Zharkov DO, Grollman AP. Catalytic mechanism of Escherichia coli endonuclease VIII: roles of the intercalation loop and the zinc finger. Biochemistry. 2006 Oct 3;45(39):12039-49. PMID:17002303 doi:10.1021/bi060663e
- ↑ Pereira de Jesus K, Serre L, Zelwer C, Castaing B. Structural insights into abasic site for Fpg specific binding and catalysis: comparative high-resolution crystallographic studies of Fpg bound to various models of abasic site analogues-containing DNA. Nucleic Acids Res. 2005 Oct 20;33(18):5936-44. Print 2005. PMID:16243784 doi:http://dx.doi.org/33/18/5936
- ↑ Sugahara M, Mikawa T, Kumasaka T, Yamamoto M, Kato R, Fukuyama K, Inoue Y, Kuramitsu S. Crystal structure of a repair enzyme of oxidatively damaged DNA, MutM (Fpg), from an extreme thermophile, Thermus thermophilus HB8. EMBO J. 2000 Aug 1;19(15):3857-69. PMID:10921868 doi:http://dx.doi.org/10.1093/emboj/19.15.3857
- ↑ Burgess S, Jaruga P, Dodson ML, Dizdaroglu M, Lloyd RS. Determination of active site residues in Escherichia coli endonuclease VIII. J Biol Chem. 2002 Jan 25;277(4):2938-44. Epub 2001 Nov 15. PMID:11711552 doi:10.1074/jbc.M110499200
- ↑ Zharkov DO, Golan G, Gilboa R, Fernandes AS, Gerchman SE, Kycia JH, Rieger RA, Grollman AP, Shoham G. Structural analysis of an Escherichia coli endonuclease VIII covalent reaction intermediate. EMBO J. 2002 Feb 15;21(4):789-800. PMID:11847126 doi:10.1093/emboj/21.4.789
- ↑ Zharkov DO, Golan G, Gilboa R, Fernandes AS, Gerchman SE, Kycia JH, Rieger RA, Grollman AP, Shoham G. Structural analysis of an Escherichia coli endonuclease VIII covalent reaction intermediate. EMBO J. 2002 Feb 15;21(4):789-800. PMID:11847126 doi:10.1093/emboj/21.4.789
- ↑ O'Connor TR, Graves RJ, de Murcia G, Castaing B, Laval J. Fpg protein of Escherichia coli is a zinc finger protein whose cysteine residues have a structural and/or functional role. J Biol Chem. 1993 Apr 25;268(12):9063-70. PMID:8473347
- ↑ Tchou J, Michaels ML, Miller JH, Grollman AP. Function of the zinc finger in Escherichia coli Fpg protein. J Biol Chem. 1993 Dec 15;268(35):26738-44. PMID:8253809
- ↑ Doublie S, Bandaru V, Bond JP, Wallace SS. The crystal structure of human endonuclease VIII-like 1 (NEIL1) reveals a zincless finger motif required for glycosylase activity. Proc Natl Acad Sci U S A. 2004 Jul 13;101(28):10284-9. Epub 2004 Jul 1. PMID:15232006 doi:10.1073/pnas.0402051101
- ↑ Kathe SD, Barrantes-Reynolds R, Jaruga P, Newton MR, Burrows CJ, Bandaru V, Dizdaroglu M, Bond JP, Wallace SS. Plant and fungal Fpg homologs are formamidopyrimidine DNA glycosylases but not 8-oxoguanine DNA glycosylases. DNA Repair (Amst). 2009 May 1;8(5):643-53. Epub 2009 Feb 12. PMID:19217358 doi:10.1016/j.dnarep.2008.12.013
- ↑ Bandaru V, Zhao X, Newton MR, Burrows CJ, Wallace SS. Human endonuclease VIII-like (NEIL) proteins in the giant DNA Mimivirus. DNA Repair (Amst). 2007 Nov;6(11):1629-41. Epub 2007 Jul 12. PMID:17627905 doi:10.1016/j.dnarep.2007.05.011
- ↑ Fromme JC, Verdine GL. DNA lesion recognition by the bacterial repair enzyme MutM. J Biol Chem. 2003 Dec 19;278(51):51543-8. Epub 2003 Oct 1. PMID:14525999 doi:10.1074/jbc.M307768200
- ↑ Doublie S, Bandaru V, Bond JP, Wallace SS. The crystal structure of human endonuclease VIII-like 1 (NEIL1) reveals a zincless finger motif required for glycosylase activity. Proc Natl Acad Sci U S A. 2004 Jul 13;101(28):10284-9. Epub 2004 Jul 1. PMID:15232006 doi:10.1073/pnas.0402051101
- ↑ Burgess S, Jaruga P, Dodson ML, Dizdaroglu M, Lloyd RS. Determination of active site residues in Escherichia coli endonuclease VIII. J Biol Chem. 2002 Jan 25;277(4):2938-44. Epub 2001 Nov 15. PMID:11711552 doi:10.1074/jbc.M110499200
- ↑ Golan G, Zharkov DO, Feinberg H, Fernandes AS, Zaika EI, Kycia JH, Grollman AP, Shoham G. Structure of the uncomplexed DNA repair enzyme endonuclease VIII indicates significant interdomain flexibility. Nucleic Acids Res. 2005 Sep 6;33(15):5006-16. Print 2005. PMID:16145054 doi:http://dx.doi.org/33/15/5006
- ↑ Zharkov DO, Golan G, Gilboa R, Fernandes AS, Gerchman SE, Kycia JH, Rieger RA, Grollman AP, Shoham G. Structural analysis of an Escherichia coli endonuclease VIII covalent reaction intermediate. EMBO J. 2002 Feb 15;21(4):789-800. PMID:11847126 doi:10.1093/emboj/21.4.789
- ↑ Burgess S, Jaruga P, Dodson ML, Dizdaroglu M, Lloyd RS. Determination of active site residues in Escherichia coli endonuclease VIII. J Biol Chem. 2002 Jan 25;277(4):2938-44. Epub 2001 Nov 15. PMID:11711552 doi:10.1074/jbc.M110499200
- ↑ Burgess S, Jaruga P, Dodson ML, Dizdaroglu M, Lloyd RS. Determination of active site residues in Escherichia coli endonuclease VIII. J Biol Chem. 2002 Jan 25;277(4):2938-44. Epub 2001 Nov 15. PMID:11711552 doi:10.1074/jbc.M110499200
- ↑ Sidorkina OM, Laval J. Role of lysine-57 in the catalytic activities of Escherichia coli formamidopyrimidine-DNA glycosylase (Fpg protein). Nucleic Acids Res. 1998 Dec 1;26(23):5351-7. PMID:9826758
- ↑ Zharkov DO, Golan G, Gilboa R, Fernandes AS, Gerchman SE, Kycia JH, Rieger RA, Grollman AP, Shoham G. Structural analysis of an Escherichia coli endonuclease VIII covalent reaction intermediate. EMBO J. 2002 Feb 15;21(4):789-800. PMID:11847126 doi:10.1093/emboj/21.4.789
- ↑ Kropachev KY, Zharkov DO, Grollman AP. Catalytic mechanism of Escherichia coli endonuclease VIII: roles of the intercalation loop and the zinc finger. Biochemistry. 2006 Oct 3;45(39):12039-49. PMID:17002303 doi:10.1021/bi060663e
- ↑ Burgess S, Jaruga P, Dodson ML, Dizdaroglu M, Lloyd RS. Determination of active site residues in Escherichia coli endonuclease VIII. J Biol Chem. 2002 Jan 25;277(4):2938-44. Epub 2001 Nov 15. PMID:11711552 doi:10.1074/jbc.M110499200
- ↑ Rabow LE, Kow YW. Mechanism of action of base release by Escherichia coli Fpg protein: role of lysine 155 in catalysis. Biochemistry. 1997 Apr 22;36(16):5084-96. PMID:9125531 doi:10.1021/bi963005a
- ↑ Rabow L, Venkataraman R, Kow YW. Mechanism of action of Escherichia coli formamidopyrimidine N-glycosylase: role of K155 in substrate binding and product release. Prog Nucleic Acid Res Mol Biol. 2001;68:223-34. PMID:11554299
- ↑ Burgess S, Jaruga P, Dodson ML, Dizdaroglu M, Lloyd RS. Determination of active site residues in Escherichia coli endonuclease VIII. J Biol Chem. 2002 Jan 25;277(4):2938-44. Epub 2001 Nov 15. PMID:11711552 doi:10.1074/jbc.M110499200
- ↑ Kropachev KY, Zharkov DO, Grollman AP. Catalytic mechanism of Escherichia coli endonuclease VIII: roles of the intercalation loop and the zinc finger. Biochemistry. 2006 Oct 3;45(39):12039-49. PMID:17002303 doi:10.1021/bi060663e
- ↑ Burgess S, Jaruga P, Dodson ML, Dizdaroglu M, Lloyd RS. Determination of active site residues in Escherichia coli endonuclease VIII. J Biol Chem. 2002 Jan 25;277(4):2938-44. Epub 2001 Nov 15. PMID:11711552 doi:10.1074/jbc.M110499200
- ↑ Zharkov DO, Golan G, Gilboa R, Fernandes AS, Gerchman SE, Kycia JH, Rieger RA, Grollman AP, Shoham G. Structural analysis of an Escherichia coli endonuclease VIII covalent reaction intermediate. EMBO J. 2002 Feb 15;21(4):789-800. PMID:11847126 doi:10.1093/emboj/21.4.789
- ↑ Golan G, Zharkov DO, Feinberg H, Fernandes AS, Zaika EI, Kycia JH, Grollman AP, Shoham G. Structure of the uncomplexed DNA repair enzyme endonuclease VIII indicates significant interdomain flexibility. Nucleic Acids Res. 2005 Sep 6;33(15):5006-16. Print 2005. PMID:16145054 doi:http://dx.doi.org/33/15/5006
- ↑ Shinmura K, Tao H, Goto M, Igarashi H, Taniguchi T, Maekawa M, Takezaki T, Sugimura H. Inactivating mutations of the human base excision repair gene NEIL1 in gastric cancer. Carcinogenesis. 2004 Dec;25(12):2311-7. Epub 2004 Aug 19. PMID:15319300 doi:10.1093/carcin/bgh267
- ↑ Tchou J, Michaels ML, Miller JH, Grollman AP. Function of the zinc finger in Escherichia coli Fpg protein. J Biol Chem. 1993 Dec 15;268(35):26738-44. PMID:8253809
- ↑ Tchou J, Michaels ML, Miller JH, Grollman AP. Function of the zinc finger in Escherichia coli Fpg protein. J Biol Chem. 1993 Dec 15;268(35):26738-44. PMID:8253809
- ↑ Kropachev KY, Zharkov DO, Grollman AP. Catalytic mechanism of Escherichia coli endonuclease VIII: roles of the intercalation loop and the zinc finger. Biochemistry. 2006 Oct 3;45(39):12039-49. PMID:17002303 doi:10.1021/bi060663e
- ↑ O'Connor TR, Graves RJ, de Murcia G, Castaing B, Laval J. Fpg protein of Escherichia coli is a zinc finger protein whose cysteine residues have a structural and/or functional role. J Biol Chem. 1993 Apr 25;268(12):9063-70. PMID:8473347