Sandbox122
From Proteopedia
| Line 9: | Line 9: | ||
==Interaction between FMN et LOV2 domain== | ==Interaction between FMN et LOV2 domain== | ||
The <scene name='Sandbox122/Fmn_ligand/1'>FMN</scene> (Flavin MonoNucleotide) is the ligand which is responsible for the light absorption. A single molecule of FMN is bound non convalently in the interior of LOV2 domain. FMN is stabilized thanks to hydrogen bonds, Van der Waals and electrostatic interactions. For example, atoms <scene name='Sandbox122/Arg_983/1'>R983</scene> and <scene name='Sandbox122/Arg_963/1'>R967</scene>(alpha C helix) create ionic bond with phosphate group of FMN. <scene name='Sandbox122/Q970/1'>Q970</scene>, <scene name='Sandbox122/N965/1'>N965</scene> , <scene name='Sandbox122/N998/1'>N988</scene>, <scene name='Sandbox122/N1008/1'>N1008</scene> (alpha A helix and beta-strand C, D and E) aminoacid make some electrostatic interaction which stabilize FMN. Some results indicate that the majority of the FMN in the LOV2 domain exist in the protonated form. Researcher propose a reaction mechanism that involves excited-state proton transfer, on the nanosecond time scale , from the sulfhydryl group of the conserved cysteine to the N5 atom of FMN. | The <scene name='Sandbox122/Fmn_ligand/1'>FMN</scene> (Flavin MonoNucleotide) is the ligand which is responsible for the light absorption. A single molecule of FMN is bound non convalently in the interior of LOV2 domain. FMN is stabilized thanks to hydrogen bonds, Van der Waals and electrostatic interactions. For example, atoms <scene name='Sandbox122/Arg_983/1'>R983</scene> and <scene name='Sandbox122/Arg_963/1'>R967</scene>(alpha C helix) create ionic bond with phosphate group of FMN. <scene name='Sandbox122/Q970/1'>Q970</scene>, <scene name='Sandbox122/N965/1'>N965</scene> , <scene name='Sandbox122/N998/1'>N988</scene>, <scene name='Sandbox122/N1008/1'>N1008</scene> (alpha A helix and beta-strand C, D and E) aminoacid make some electrostatic interaction which stabilize FMN. Some results indicate that the majority of the FMN in the LOV2 domain exist in the protonated form. Researcher propose a reaction mechanism that involves excited-state proton transfer, on the nanosecond time scale , from the sulfhydryl group of the conserved cysteine to the N5 atom of FMN. | ||
| - | Blue light arrives on the only LOV2 <scene name='Sandbox122/Cystein_residue/1'>cystein residue</scene>(situate 4.2A from atom C(4a)) and induces formation of covalent cysteinyl-C(4a)adduct. (To explain that, refer to the scheme). This complex may interact directly with the kinase and regulate is activity. | + | Blue light arrives on the only LOV2 <scene name='Sandbox122/Cystein_residue/1'>cystein residue</scene>(situate 4.2A from atom C(4a)) and induces formation of covalent cysteinyl-C(4a)adduct. (To explain that, refer to the scheme). Somes studies showed that temperature have an influence on that adduct. In fact, at low temperature, the microenvironment determine the reactivity of the S-H group of Cys966. This complex may interact directly with the kinase and regulate is activity. |
Kinase will permit the autophosphorylation of phototropin. The rate of phosphorylatide phototropin acts to differential lateral gradients of auxin which is responsible of phototropism phenomenon. | Kinase will permit the autophosphorylation of phototropin. The rate of phosphorylatide phototropin acts to differential lateral gradients of auxin which is responsible of phototropism phenomenon. | ||
[[Image:1g28.jpg | thumb]] | [[Image:1g28.jpg | thumb]] | ||
[[Image:1G28.pdb_b.jpg | thumb]] | [[Image:1G28.pdb_b.jpg | thumb]] | ||
Revision as of 12:47, 10 November 2009
|
Phototropin is a blue light receptor involved in the phototropism (it is a phenomenon which is growth directed by light). This protein is a dimer (phot1 and phot2) and each subunit contains three domains : LOV1, LOV2 (light,oxygen or voltage) domains and a kinase domain.
LOV2 domain was, here, chosen for study.
Structure of LOV2
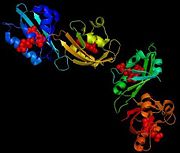
The structure of a flaving domain, LOV2, from the chimeric phototropin photoreceptor PHY3, has been determined thanks to X-Ray diffraction. This domain is composed by four chains which are identical A, B, C and D. It's a L-polypeptide which weight 50552,99 Da. Each chains has 104 aminoacids and possess four (28 residues which represents 26%) and six B (37 residues which represent 35%).
Interaction between FMN et LOV2 domain
The (Flavin MonoNucleotide) is the ligand which is responsible for the light absorption. A single molecule of FMN is bound non convalently in the interior of LOV2 domain. FMN is stabilized thanks to hydrogen bonds, Van der Waals and electrostatic interactions. For example, atoms and (alpha C helix) create ionic bond with phosphate group of FMN. , , , (alpha A helix and beta-strand C, D and E) aminoacid make some electrostatic interaction which stabilize FMN. Some results indicate that the majority of the FMN in the LOV2 domain exist in the protonated form. Researcher propose a reaction mechanism that involves excited-state proton transfer, on the nanosecond time scale , from the sulfhydryl group of the conserved cysteine to the N5 atom of FMN. Blue light arrives on the only LOV2 (situate 4.2A from atom C(4a)) and induces formation of covalent cysteinyl-C(4a)adduct. (To explain that, refer to the scheme). Somes studies showed that temperature have an influence on that adduct. In fact, at low temperature, the microenvironment determine the reactivity of the S-H group of Cys966. This complex may interact directly with the kinase and regulate is activity. Kinase will permit the autophosphorylation of phototropin. The rate of phosphorylatide phototropin acts to differential lateral gradients of auxin which is responsible of phototropism phenomenon.