T4 RNA ligase 2 (Rnl2)
From Proteopedia
| Line 12: | Line 12: | ||
Although this protein is designed to counteract the bacterial host’s defence mechanisms against the virus, its activity as a ligase is analogous to tRNA splicing (introns removed from anticodon loop) and RNA editing by several different kinds of RNA editing ligases (REL)s. Therefore it have been grouped into a subfamily of RNA ligases including REL-1 which is essential for the survival of Trypanosoma, a monophyletic group of unicellular paracitic flagella protozoa that causes sleeping sickness. Another commonality shared between RNA ligases as well as DNA ligases and capping enzymes is the fact that the mechanism by which they function involves the formation of a covalent enzyme-(lysyl-N)-NMP intermediate prior to the formation of the phophodiester bond between 5’ and 3’ ends of RNA strands. Hence these groups have been classified as a super family of enzymes, characteristic of the above intermediate. | Although this protein is designed to counteract the bacterial host’s defence mechanisms against the virus, its activity as a ligase is analogous to tRNA splicing (introns removed from anticodon loop) and RNA editing by several different kinds of RNA editing ligases (REL)s. Therefore it have been grouped into a subfamily of RNA ligases including REL-1 which is essential for the survival of Trypanosoma, a monophyletic group of unicellular paracitic flagella protozoa that causes sleeping sickness. Another commonality shared between RNA ligases as well as DNA ligases and capping enzymes is the fact that the mechanism by which they function involves the formation of a covalent enzyme-(lysyl-N)-NMP intermediate prior to the formation of the phophodiester bond between 5’ and 3’ ends of RNA strands. Hence these groups have been classified as a super family of enzymes, characteristic of the above intermediate. | ||
| - | The mechanism for the formation of this intermediate and the phosphodiester bond resulting in RNA strand repair is outlined below. First ATP reacts with the active site of Rln2. Adenosine mono phosphate <scene name='Sandbox_157/T4_rnl2_active_site/3'>(AMP)</scene> binds to the <scene name='Sandbox_157/T4_rnl2_active_site/1'>active site (light green)</scene> accompanied by the release of inorganic phosphate which provides the energy. The adenylate binds at the bottom of the <scene name='Sandbox_157/T4_rnl2_active_site/1'>active site</scene> (motif 1) where it is squished between the aromatic rings of <scene name='Sandbox_157/T4_rnl2_active_site/4'>Phe119 (light blue)</scene> of motif IIIa, <scene name='Sandbox_157/T4_rnl2_active_site/5'>Val 207 (purple)</scene> of motif III and <scene name='Sandbox_157/T4_rnl2_active_site/6'>Lys 35 (yellow)</scene> of motif I. <scene name='Sandbox_157/T4_rnl2_active_site/14'>Lys225 (orange)</scene> and <scene name='Sandbox_157/T4_rnl2_active_site/15'>Lys227 (peach)</scene> of motif V, which are essential for Rln2 activity, are also involved in coordinating the AMP phosphate after binding. During ligase adenylation the alpha phosphate of ATP is thought to be stabilized by Lys 225 and 227 prior to release of the other phosphate groups. The specificity of Rln2 for ATP as opposed to other NTP substrates in this step are thought to be attributed to hydrogen bonding between backbone residues and the adenine base. Such bonds occur between:<scene name='Sandbox_157/T4_rnl2_active_site/7'>N7 (on AMP) and the backbone amide of Ile36 (dark green),</scene> <scene name='Sandbox_157/T4_rnl2_active_site/8'>the exocyclic 6-amino group (N6) and main chain carbonyl (O) of Glu34 (dark purple),</scene> <scene name='Sandbox_157/T4_rnl2_active_site/9'>N1 and Lys209 (dark red) (water (white circles) mediated)</scene>. After Binding AMP, the Rnl2 transfers AMP to the 5' phosphate of the RNA strand. The binding of Rnl2 to RNA, required for this step, can be partially attributed to the electrostatic structure of the enzyme. Since the active site surface surounding AMP is possitively charged it may contribute to the affinity of the enzyme for negatively charge 5' phosphate of pRNA. Note that all interactions between pRNA and Rnl2 also require the essential C-terminal domain which is not shown in the crystalized structure here. This may be due to the essential positioning requirements for the placement of ATP in correct close proximity and orientation to the 5' phosphate for the next step (Figure 1). [[Image:Rnl2 Electorstatic potential.jpg | thumb|alt=T4 Rnl2.| Figure 1. Electrostatic potential of Rnl2 where blue represents positive charges and red represents net negative charged regions. Majority of the surfaces have a net negative charge while positve charges are localized around AMP in the surrounding active site.]] The interactions between the ribose sugar and the protein back bone are all thought to contribute to this adenyltransferase function as well as overall pRNA ligation, where the interaction between the<scene name='Sandbox_157/T4_rnl2_active_site/16'> 2' oxygen of the ribose sugar and Glu99 (teal)</scene> is thought to be most essential to this step. As well the <scene name='Sandbox_157/T4_rnl2_active_site/17'>Arg55 (dark blue) guanidinium group (NH2, NE, NH1)</scene>, which coordinates the 3' O of the ribose sugar, is thought to interact with the 5'-PO<sub>4</sub> of RNA during this adenylation step as well as during initial adenylation of the enzyme by ATP (Figure 2). Likewise Glu204 has been found to be essential to both of these steps as well. The <scene name='Sandbox_157/T4_rnl2_active_site/10'>π stacking between Phe119 and the adenylate base</scene> is thought to be critical for the next step; transfer of the phosphate from the 5’ end of the RNA strand to the 3’ OH end of the RNA strand to form the phosphodiester bond Figure 2. The actual catalytic action of this step however, is thought to be carried out by <scene name='Sandbox_157/T4_rnl2_active_site/18'> | + | The mechanism for the formation of this intermediate and the phosphodiester bond resulting in RNA strand repair is outlined below. First ATP reacts with the active site of Rln2. Adenosine mono phosphate <scene name='Sandbox_157/T4_rnl2_active_site/3'>(AMP)</scene> binds to the <scene name='Sandbox_157/T4_rnl2_active_site/1'>active site (light green)</scene> accompanied by the release of inorganic phosphate which provides the energy. The adenylate binds at the bottom of the <scene name='Sandbox_157/T4_rnl2_active_site/1'>active site</scene> (motif 1) where it is squished between the aromatic rings of <scene name='Sandbox_157/T4_rnl2_active_site/4'>Phe119 (light blue)</scene> of motif IIIa, <scene name='Sandbox_157/T4_rnl2_active_site/5'>Val 207 (purple)</scene> of motif III and <scene name='Sandbox_157/T4_rnl2_active_site/6'>Lys 35 (yellow)</scene> of motif I. <scene name='Sandbox_157/T4_rnl2_active_site/14'>Lys225 (orange)</scene> and <scene name='Sandbox_157/T4_rnl2_active_site/15'>Lys227 (peach)</scene> of motif V, which are essential for Rln2 activity, are also involved in coordinating the AMP phosphate after binding. During ligase adenylation the alpha phosphate of ATP is thought to be stabilized by Lys 225 and 227 prior to release of the other phosphate groups. The specificity of Rln2 for ATP as opposed to other NTP substrates in this step are thought to be attributed to hydrogen bonding between backbone residues and the adenine base. Such bonds occur between:<scene name='Sandbox_157/T4_rnl2_active_site/7'>N7 (on AMP) and the backbone amide of Ile36 (dark green),</scene> <scene name='Sandbox_157/T4_rnl2_active_site/8'>the exocyclic 6-amino group (N6) and main chain carbonyl (O) of Glu34 (dark purple),</scene> <scene name='Sandbox_157/T4_rnl2_active_site/9'>N1 and Lys209 (dark red) (water (white circles) mediated)</scene>. After Binding AMP, the Rnl2 transfers AMP to the 5' phosphate of the RNA strand. The binding of Rnl2 to RNA, required for this step, can be partially attributed to the electrostatic structure of the enzyme. Since the active site surface surounding AMP is possitively charged it may contribute to the affinity of the enzyme for negatively charge 5' phosphate of pRNA. Note that all interactions between pRNA and Rnl2 also require the essential C-terminal domain which is not shown in the crystalized structure here. This may be due to the essential positioning requirements for the placement of ATP in correct close proximity and orientation to the 5' phosphate for the next step (Figure 1). [[Image:Rnl2 Electorstatic potential.jpg | thumb|alt=T4 Rnl2.| Figure 1. Electrostatic potential of Rnl2 where blue represents positive charges and red represents net negative charged regions. Majority of the surfaces have a net negative charge while positve charges are localized around AMP in the surrounding active site.]] The interactions between the ribose sugar and the protein back bone are all thought to contribute to this adenyltransferase function as well as overall pRNA ligation, where the interaction between the<scene name='Sandbox_157/T4_rnl2_active_site/16'> 2' oxygen of the ribose sugar and Glu99 (teal)</scene> is thought to be most essential to this step. As well the <scene name='Sandbox_157/T4_rnl2_active_site/17'>Arg55 (dark blue) guanidinium group (NH2, NE, NH1)</scene>, which coordinates the 3' O of the ribose sugar, is thought to interact with the 5'-PO<sub>4</sub> of RNA during this adenylation step as well as during initial adenylation of the enzyme by ATP (Figure 2). Likewise Glu204 has been found to be essential to both of these steps as well. The <scene name='Sandbox_157/T4_rnl2_active_site/10'>π stacking between Phe119 and the adenylate base</scene> is thought to be critical for the next step; transfer of the phosphate from the 5’ end of the RNA strand to the 3’ OH end of the RNA strand to form the phosphodiester bond Figure 2. The actual catalytic action of this step however, is thought to be carried out by <scene name='Sandbox_157/T4_rnl2_active_site/18'>His37 (light green)</scene> in motif 1. Ho et al. hypothesized that His37 sets the boundaries of the binding site for the 3' OH RNA strand which acts as a nucleophile in this step and hence is necessary for catalyzing this step. |
| + | ---- | ||
| + | Structural components contributing to the active site: | ||
---- | ---- | ||
| - | |||
Current Refences: Ho et al. 2004 Structure and Mechanism of RNA Ligase. 12:327-339 | Current Refences: Ho et al. 2004 Structure and Mechanism of RNA Ligase. 12:327-339 | ||
Revision as of 01:54, 18 February 2010
|
Overview:
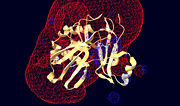
T4 RNA ligase 2 (Rnl2) (1-249) is a T4 bacteriophage ligase, which functions to counter bacterial defence by repairing broken anticodon loops of tRNA coding for lysine, caused by PrrC, which is nessessary for viral replication and proliferation. Note that the structure shown is based on the truncated enzyme form (1-249) where the total chain length of Rnl2 is 334 residues. The overall structure, at 100K, pH 8.5, consists of a single chain polymer (233 residues + 15 His tag residues) composed of 2, 6 antiparalel stranded, beta sheets and 7 alpha helices. The structure contains an adenosine monophosphate ligand in its active site. The enzyme contains 5 nucleotidyl transferase motifs, (green, blue, pink, light blue, red) which are all involved either directly for indirectly with the function of the active site.
Protein function:
Although this protein is designed to counteract the bacterial host’s defence mechanisms against the virus, its activity as a ligase is analogous to tRNA splicing (introns removed from anticodon loop) and RNA editing by several different kinds of RNA editing ligases (REL)s. Therefore it have been grouped into a subfamily of RNA ligases including REL-1 which is essential for the survival of Trypanosoma, a monophyletic group of unicellular paracitic flagella protozoa that causes sleeping sickness. Another commonality shared between RNA ligases as well as DNA ligases and capping enzymes is the fact that the mechanism by which they function involves the formation of a covalent enzyme-(lysyl-N)-NMP intermediate prior to the formation of the phophodiester bond between 5’ and 3’ ends of RNA strands. Hence these groups have been classified as a super family of enzymes, characteristic of the above intermediate.
The mechanism for the formation of this intermediate and the phosphodiester bond resulting in RNA strand repair is outlined below. First ATP reacts with the active site of Rln2. Adenosine mono phosphate binds to the accompanied by the release of inorganic phosphate which provides the energy. The adenylate binds at the bottom of the (motif 1) where it is squished between the aromatic rings of of motif IIIa, of motif III and of motif I. and of motif V, which are essential for Rln2 activity, are also involved in coordinating the AMP phosphate after binding. During ligase adenylation the alpha phosphate of ATP is thought to be stabilized by Lys 225 and 227 prior to release of the other phosphate groups. The specificity of Rln2 for ATP as opposed to other NTP substrates in this step are thought to be attributed to hydrogen bonding between backbone residues and the adenine base. Such bonds occur between: . After Binding AMP, the Rnl2 transfers AMP to the 5' phosphate of the RNA strand. The binding of Rnl2 to RNA, required for this step, can be partially attributed to the electrostatic structure of the enzyme. Since the active site surface surounding AMP is possitively charged it may contribute to the affinity of the enzyme for negatively charge 5' phosphate of pRNA. Note that all interactions between pRNA and Rnl2 also require the essential C-terminal domain which is not shown in the crystalized structure here. This may be due to the essential positioning requirements for the placement of ATP in correct close proximity and orientation to the 5' phosphate for the next step (Figure 1).Structural components contributing to the active site:
Current Refences: Ho et al. 2004 Structure and Mechanism of RNA Ligase. 12:327-339
Proteopedia Page Contributors and Editors (what is this?)
William Eisbrenner, Alexander Berchansky, David Canner, Sonja Senekovic, Michal Harel, Andrea Gorrell