Phosphoglucoisomerase
From Proteopedia
(→'''Structure''') |
(→'''Structure''') |
||
| Line 11: | Line 11: | ||
=='''Structure'''== | =='''Structure'''== | ||
<applet load="1hox" size="300" color="white" frame="true" align="right" caption="Phosphoglucose isomerase" /> | <applet load="1hox" size="300" color="white" frame="true" align="right" caption="Phosphoglucose isomerase" /> | ||
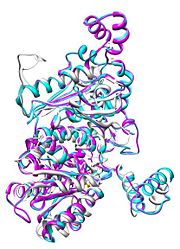
| - | [[Image:Align.jpg|thumb|left|''' | + | [[Image:Align.jpg|thumb|left|'''Figure 1.''' Multiple alignment PGI - ''Geobacillus stearothermophilus'' (white),'' Homo sapiens'' (pink), ''Oryctolagus cuniculus'' (turquoise)]] |
Phosphoglucose isomerase exists in the cell usually as a <scene name='Stancu_Phosphoglucoisomerase_Sandbox_1/Dimer/1'>homodimer</scene>, nevertheless outside of the cell, it has been isolated as <scene name='Stancu_Phosphoglucoisomerase_Sandbox_1/Monomer/1'>monomeric</scene> structure. PGI has essentially an identical fold in all of the characterized species (see '''Figure 1'''). The <scene name='Stancu_Phosphoglucoisomerase_Sandbox_1/Sec_struct/1'>secondary structure</scene> of phosphoglucose isomerase is charaterized by an αβα conformation, on each of its two domains. The smaller domain is characterized by 5 parallel β-sheets, while the larger domain if formed out of 6 parallel/antiparallel β-sheets. Furthermore, another charateristical trait is a residue extension at the C-terminus, which wraps around the other monomer in the dimeric conformation | Phosphoglucose isomerase exists in the cell usually as a <scene name='Stancu_Phosphoglucoisomerase_Sandbox_1/Dimer/1'>homodimer</scene>, nevertheless outside of the cell, it has been isolated as <scene name='Stancu_Phosphoglucoisomerase_Sandbox_1/Monomer/1'>monomeric</scene> structure. PGI has essentially an identical fold in all of the characterized species (see '''Figure 1'''). The <scene name='Stancu_Phosphoglucoisomerase_Sandbox_1/Sec_struct/1'>secondary structure</scene> of phosphoglucose isomerase is charaterized by an αβα conformation, on each of its two domains. The smaller domain is characterized by 5 parallel β-sheets, while the larger domain if formed out of 6 parallel/antiparallel β-sheets. Furthermore, another charateristical trait is a residue extension at the C-terminus, which wraps around the other monomer in the dimeric conformation | ||
| Line 20: | Line 20: | ||
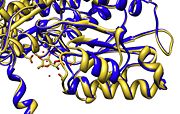
Another characteristic of phosphoglucose isomerase is that binding of substrate at the active site, induces a movement in the conformation of the enzyme. This can be seen in Figure 2 as change in the position of an α helix. | Another characteristic of phosphoglucose isomerase is that binding of substrate at the active site, induces a movement in the conformation of the enzyme. This can be seen in Figure 2 as change in the position of an α helix. | ||
| - | [[Image:Active_site_movement.jpg|thumb|right|''' | + | [[Image:Active_site_movement.jpg|thumb|right|'''Figure 2.''' Substrate induced movement]] |
=='''Mechanism'''== | =='''Mechanism'''== | ||
Revision as of 03:17, 1 March 2010
|
Phosphoglucoisomerase (alternatively known as phosphoglucose isomerase or Glucose-6-phosphate isomerase) are a group of enzymes of the isomerase family (EC 5.3.1.9), so named for their main function in glycolysis and gluconeogenesis. In both these pathways phosphoglucose isomerase (PGI) is used to inter-convert glucose-6-phosphate and fructose 6-phosphate, reaction driven by the relative concentrations of these sugars in the cytoplasmic matrix of the cell.
Phosphoglucoisomerase is also know for a list of activities outside the cells:
- Neuroleukin (NLK)- nerve growth factor secreted by T cells, also used to stimulate the production of immunoglobulin [1].
- Autocrine motility factor (AMF)- product of tumor cells, it promotes cell migration and viewed as a possible cause in cancer metastasis[2].
- Maturation factor(MF) [3]
- Myofibril-bound serine protese inhibitor (MBSPI)[4]
Contents |
Structure
|
Phosphoglucose isomerase exists in the cell usually as a , nevertheless outside of the cell, it has been isolated as structure. PGI has essentially an identical fold in all of the characterized species (see Figure 1). The of phosphoglucose isomerase is charaterized by an αβα conformation, on each of its two domains. The smaller domain is characterized by 5 parallel β-sheets, while the larger domain if formed out of 6 parallel/antiparallel β-sheets. Furthermore, another charateristical trait is a residue extension at the C-terminus, which wraps around the other monomer in the dimeric conformation
Phosphoglucose isomerase has a molecular mass of proximately 55 kDa.
Active Site - Mammalian PGI shows a degree of ( dark red represent highly conserved regions - dark blue regions that are variable) of about 90 %. The is the region with highest observed conservation, with a number of residues that are crucial in the mechanism of the enzyme (Lys210, Gln353, Glu357, Gln511, Lys518, His388b).
Another characteristic of phosphoglucose isomerase is that binding of substrate at the active site, induces a movement in the conformation of the enzyme. This can be seen in Figure 2 as change in the position of an α helix.
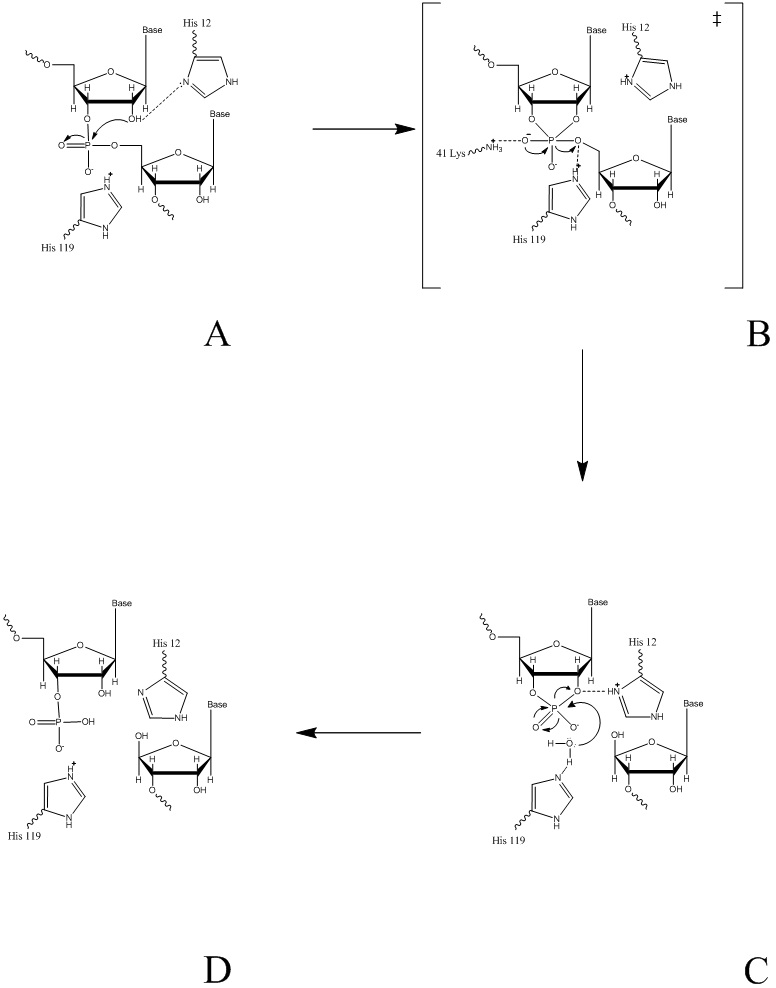
Mechanism
The proposed reaction mechanism of PGI for the conversion of glucose-6-phosphate to fructose 6-phosphate involves an acid/base catalysis by the enzyme.
Links
References
- ↑ Gurney ME, Heinrich SP, Lee MR, Yin HS. Molecular cloning and expression of neuroleukin, a neurotrophic factor for spinal and sensory neurons. Science. 1986 Oct 31;234(4776):566-74. PMID:3764429
- ↑ Tanaka N, Haga A, Uemura H, Akiyama H, Funasaka T, Nagase H, Raz A, Nakamura KT. Inhibition mechanism of cytokine activity of human autocrine motility factor examined by crystal structure analyses and site-directed mutagenesis studies. J Mol Biol. 2002 May 10;318(4):985-97. PMID:12054796 doi:10.1016/S0022-2836(02)00186-9
- ↑ Xu W, Seiter K, Feldman E, Ahmed T, Chiao JW. The differentiation and maturation mediator for human myeloid leukemia cells shares homology with neuroleukin or phosphoglucose isomerase. Blood. 1996 Jun 1;87(11):4502-6. PMID:8639816
- ↑ Cao MJ, Osatomi K, Matsuda R, Ohkubo M, Hara K, Ishihara T. Purification of a novel serine proteinase inhibitor from the skeletal muscle of white croaker (Argyrosomus argentatus). Biochem Biophys Res Commun. 2000 Jun 7;272(2):485-9. PMID:10833440 doi:10.1006/bbrc.2000.2803
- ↑ Read J, Pearce J, Li X, Muirhead H, Chirgwin J, Davies C. The crystal structure of human phosphoglucose isomerase at 1.6 A resolution: implications for catalytic mechanism, cytokine activity and haemolytic anaemia. J Mol Biol. 2001 Jun 1;309(2):447-63. PMID:11371164 doi:10.1006/jmbi.2001.4680
- ↑ Lee JH, Chang KZ, Patel V, Jeffery CJ. Crystal structure of rabbit phosphoglucose isomerase complexed with its substrate D-fructose 6-phosphate. Biochemistry. 2001 Jul 3;40(26):7799-805. PMID:11425306
Proteopedia Page Contributors and Editors (what is this?)
Joel L. Sussman, Bogdan Stancu, Michal Harel, Andrew Gilman, Alexander Berchansky, David Canner