Inositol 1,4,5-Trisphosphate Receptor
From Proteopedia
| Line 35: | Line 35: | ||
Highly basic amino acid residues are present on both domains and are responsile for the binding of InsP<sub>3</sub> to InsP<sub>3</sub>R.<sup>[1]</sup> In binding, water molecules are involved in hydrogen bonding between InsP<sub>3</sub> and its receptor as well as interactions between protein side chains and phosphorous.<sup>[1]</sup> Coordination of phosphorous groups is mediated by residues in both the β-domain and α-domain. The hydroxyl groups of InsP<sub>3</sub> play a small role in binding to InsP<sub>3</sub>.<sup>[1]</sup> Additionally, 9 out of 12 Arg/Lys residues play a very important role in ligand binding and salt bridges to stabilize between the domain regions.<sup>[1]</sup> The non-basic residues T266, T267, G268, and Y567 are also integral in Insp<sub>3</sub> coordination: if T267, G268 or Y567 residues are mutated then there will be a significant reduction in ligand binding.<sup>[1]</sup> | Highly basic amino acid residues are present on both domains and are responsile for the binding of InsP<sub>3</sub> to InsP<sub>3</sub>R.<sup>[1]</sup> In binding, water molecules are involved in hydrogen bonding between InsP<sub>3</sub> and its receptor as well as interactions between protein side chains and phosphorous.<sup>[1]</sup> Coordination of phosphorous groups is mediated by residues in both the β-domain and α-domain. The hydroxyl groups of InsP<sub>3</sub> play a small role in binding to InsP<sub>3</sub>.<sup>[1]</sup> Additionally, 9 out of 12 Arg/Lys residues play a very important role in ligand binding and salt bridges to stabilize between the domain regions.<sup>[1]</sup> The non-basic residues T266, T267, G268, and Y567 are also integral in Insp<sub>3</sub> coordination: if T267, G268 or Y567 residues are mutated then there will be a significant reduction in ligand binding.<sup>[1]</sup> | ||
| + | |||
| + | |||
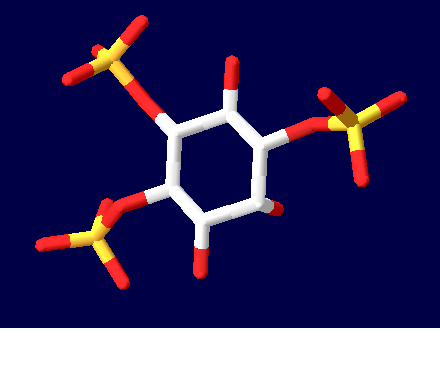
| + | [[Image:Ligand1.PNG]] | ||
| + | A representation of the InsP<sub>3</sub>R ligand, InsP<sub>3</sub> | ||
| + | |||
== Function == | == Function == | ||
---- | ---- | ||
Revision as of 00:23, 18 March 2010
| Please do NOT make changes to this Sandbox until after April 23, 2010. Sandboxes 151-200 are reserved until then for use by the Chemistry 307 class at UNBC taught by Prof. Andrea Gorrell. |
|
Inositol 1,4,5-trisphosphate receptor binding protein is a ubiquitous protein involved in the Ca2+ signalling processes in a variety of organisms [1]
Contents |
Overall Structure
The specific type of inositol 1,4,5-trisphosphate receptor (InsP3R) protein discussed here is the mouse type one InsP3R, also called InsP3R1. This polypeptide contains three major regions: the amino terminal inositol 1,4,5-trisphosphate (InsP3) binding region, the central modulatory region, and the carboxy-terminus channel region.1 The protein forms an L-shaped structure composed of two asymmetric domains perpendicular to each other.[1] The N-terminal domain is made up of 12 β-strands and 2 single-turn helices, which come together to form a barrel.[1] The C-terminal end is quite different, consisting of a bundle made of eight α-helices.[1] The interface of the two domains is lined with basic residues and forms the receptor site for InsP3.[1] The overall structure with the ligand bound can be seen here:
Domain Structure
The protein fold of the β-domain can also be called the β-trefoil. This element is present in other proteins as well, including fibroblast growth factors and mannose receptors.[1] In the case of the InsP3R β-trefoil, the structure was found to be very similar to the β-trefoil of the mannose receptor. In the β-domain of InsP3R1, three of six two-stranded hairpins come together to form a barrel and the other three form a triangular cap for the barrel.[1]
The α-domain of InsP3R shows a high degree of homology with an element called an armidillo repeat fold found in proteins such as β-catenin and importins.[1] In β-catenin and importins, the armadillo repeat functions as a motif for protein-protein interactions.[1] Within the α-domain of mouse InsP3R1, there are two large, highly conserved surfaces.[1] Both regions are rich in aromatic residues, indicating that they may function as interaction sites for parts of the receptor or other cellular proteins.[1] A possible option for this kind of binding domain would be the InsP3 binding suppressor domain present at the N-terminus which reduces the binding affinity for the InsP3 ligand.[1] The structures of the two domains can be seen in the image below.
Binding the InsP3 Ligand
Highly basic amino acid residues are present on both domains and are responsile for the binding of InsP3 to InsP3R.[1] In binding, water molecules are involved in hydrogen bonding between InsP3 and its receptor as well as interactions between protein side chains and phosphorous.[1] Coordination of phosphorous groups is mediated by residues in both the β-domain and α-domain. The hydroxyl groups of InsP3 play a small role in binding to InsP3.[1] Additionally, 9 out of 12 Arg/Lys residues play a very important role in ligand binding and salt bridges to stabilize between the domain regions.[1] The non-basic residues T266, T267, G268, and Y567 are also integral in Insp3 coordination: if T267, G268 or Y567 residues are mutated then there will be a significant reduction in ligand binding.[1]
Function
Proteopedia Page Contributors and Editors (what is this?)
Shannon King, Alexander Berchansky, Michal Harel, Ann Taylor, David Canner, Andrea Gorrell, Jaclyn Gordon
