Sandbox 181
From Proteopedia
| Line 3: | Line 3: | ||
=Structure= | =Structure= | ||
| - | Glutathione reductase belongs to the larger family of [[flavoezymes]], which use a [[flavin adenine dinucleotide]] (FAD) or [[flavin mononucleotide]] (FMN) in catalysis. It is | + | Glutathione reductase belongs to the larger family of [[flavoezymes]], which use a [[flavin adenine dinucleotide]] (FAD) or [[flavin mononucleotide]] (FMN) in catalysis. It is a disulfide oxiodreductase homodimer of 52kD monomers of which, each has three domains: |
(<scene name='Sandbox_181/Fad_ndp_highlight/1'>FAD and NADPH Highlighted</scene>) | (<scene name='Sandbox_181/Fad_ndp_highlight/1'>FAD and NADPH Highlighted</scene>) | ||
1. NADPH-binding domain (yellow), | 1. NADPH-binding domain (yellow), | ||
Revision as of 20:27, 19 March 2010
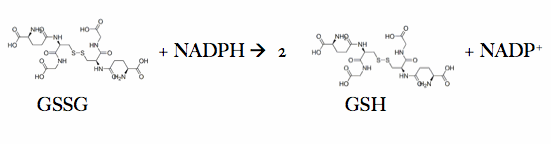
Glutathione reductase, also known as GSH reductase, is found in the human cells and converts oxidized glutathione (GSSG) to two molecules of reduced gluthatione (GSH).
| |||||||
| 3djj, resolution 1.10Å () | |||||||
|---|---|---|---|---|---|---|---|
| Ligands: | , , , , | ||||||
| Gene: | GSR, GLUR, GRD1 (HUMAN) | ||||||
| Activity: | Glutathione-disulfide reductase, with EC number 1.8.1.7 | ||||||
| Related: | 3djg | ||||||
| |||||||
| |||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
Contents |
Structure
Glutathione reductase belongs to the larger family of flavoezymes, which use a flavin adenine dinucleotide (FAD) or flavin mononucleotide (FMN) in catalysis. It is a disulfide oxiodreductase homodimer of 52kD monomers of which, each has three domains: () 1. NADPH-binding domain (yellow), 2. FAD-binding domain (red), 3. dimerization domain.
Domain structure
Reaction
The action of glutathione reductase proceeds through a cyclic series of structures in differing redox states . NADPH binds causing a transient reduction of flavin and this reduced flavin consequently reduces Cys58-Cys63 disulfide bond, forming a short lived covalent intermediate with Cys63. Following this, a stable charge-transfer complex between flavin and the Cys63 thiolate forms. After formation the NADP+ dissociates and is replaced by another NADPH. This is the end of the reductive first half of the mechanism and the oxidative half is initiated upon the binding of GSSG. The Cys58 in glutathione reductase attacks CysI of the GSSG to form a mixed disulfide between the first GS and Cys58. The second GSH is the free to leave and the disulfide bond is reformed between Cys58 and Cys63 of glutathione reductase. Finally, the first molecule of GSH is released. The glutathione reductase is then able to be recycled to allow for the binding of NADPH once again[1].
Mechanism
Function
For proper functioning and prevention of damage to a cell, GSH plays an essential role in preventing oxidative stress in human cells. GSH directly scavenges hydroxyl radicals and singlet oxygens, plays a role as a cofactor in several detoxifying enzymes, participates in amino acid transport through the plasma membrane, and can regenerate important antioxidants such as Vitamins E and C to their reactive forms [2]. The antioxidant capacity of glutathione is linked to the redox state of GSSG/2GSH inside the cell[3]. The function of GSH reductase is to maintain this narrow redox state of high reduced to oxidized ratio of GSH in the cell[4].
Consequences of a Mutation
Ultimately, a mutation in the single-copy gene coding for GSH reductase affecting its activity would disrupt the redox state of the GSSG/2GSH in the cell. If GSH were unable to be regenerated from GSSG the cellular environment would become more oxidising, a phenomenon shown to be associated will the onset of cellular apoptosis at a moderate oxidizing environment and necrosis at higher oxidizing cellular environments[5]. Elevated GSH levels (ie. a more reducing environment) stimulates cellular proliferation [6].
References
- ↑ Berkholz DS, Faber HR, Savvides SN, Karplus PA. Catalytic cycle of human glutathione reductase near 1 A resolution. J Mol Biol. 2008 Oct 3;382(2):371-84. Epub 2008 Jul 7. PMID:18638483 doi:10.1016/j.jmb.2008.06.083
- ↑ Masella R, Di Benedetto R, Vari R, Filesi C, Giovannini C. Novel mechanisms of natural antioxidant compounds in biological systems: involvement of glutathione and glutathione-related enzymes. J Nutr Biochem. 2005 Oct;16(10):577-86. PMID:16111877 doi:10.1016/j.jnutbio.2005.05.013
- ↑ Pastore A, Federici G, Bertini E, Piemonte F. Analysis of glutathione: implication in redox and detoxification. Clin Chim Acta. 2003 Jul 1;333(1):19-39. PMID:12809732
- ↑ Masella R, Di Benedetto R, Vari R, Filesi C, Giovannini C. Novel mechanisms of natural antioxidant compounds in biological systems: involvement of glutathione and glutathione-related enzymes. J Nutr Biochem. 2005 Oct;16(10):577-86. PMID:16111877 doi:10.1016/j.jnutbio.2005.05.013
- ↑ Voehringer DW, Hirschberg DL, Xiao J, Lu Q, Roederer M, Lock CB, Herzenberg LA, Steinman L, Herzenberg LA. Gene microarray identification of redox and mitochondrial elements that control resistance or sensitivity to apoptosis. Proc Natl Acad Sci U S A. 2000 Mar 14;97(6):2680-5. PMID:10716996
- ↑ Valko M, Leibfritz D, Moncol J, Cronin MT, Mazur M, Telser J. Free radicals and antioxidants in normal physiological functions and human disease. Int J Biochem Cell Biol. 2007;39(1):44-84. Epub 2006 Aug 4. PMID:16978905 doi:10.1016/j.biocel.2006.07.001
Human Glutathione Reductase
| Please do NOT make changes to this Sandbox until after April 23, 2010. Sandboxes 151-200 are reserved until then for use by the Chemistry 307 class at UNBC taught by Prof. Andrea Gorrell. |