Copper, Zinc Superoxide Dismutase
From Proteopedia
| Line 3: | Line 3: | ||
== Introduction == | == Introduction == | ||
[[Image:Superoxide_dismutase.png|thumb|Cu/Zn Superoxide Dismutase]] | [[Image:Superoxide_dismutase.png|thumb|Cu/Zn Superoxide Dismutase]] | ||
| - | Oxygen is vital to sustain life; our cells cannot live without it. Oxygen is the final acceptor of electrons in respiration, allowing us to produce more energy from food. But oxygen is also a dangerous compound (1). Reactive forms of oxygen, such as superoxide, leak from the respiratory chain and wreak havoc on the cell. Superoxide is a free radical; radicals are molecules that readily accept electrons, which make them highly reactive. They can strip electrons from proteins, lipids, or nucleic acids, thereby destroying their functions and resulting in cell dysfunction or death. Free-radical damage has been implicated in Amyotrophic lateral sclerosis (ALS), | + | Oxygen is vital to sustain life; our cells cannot live without it. Oxygen is the final acceptor of electrons in respiration at the |
| + | [http://en.wikipedia.org/wiki/Electron_transport_chain electron transport chain], allowing us to produce more energy from food. But oxygen is also a dangerous compound (1). Reactive forms of oxygen, such as superoxide, leak from the respiratory chain and wreak havoc on the cell. Superoxide is a free radical; radicals are molecules that readily accept electrons, which make them highly reactive. They can strip electrons from proteins, lipids, or nucleic acids, thereby destroying their functions and resulting in cell dysfunction or death. Free-radical damage has been implicated in [http://en.wikipedia.org/wiki/Amyotrophic_lateral_sclerosis Amyotrophic lateral sclerosis] (ALS), [http://en.wikipedia.org/wiki/Arteriosclerosis Arteriosclerosis], [http://en.wikipedia.org/wiki/Arthritis Arthritis], [http://en.wikipedia.org/wiki/Cancer Cancer], and [http://en.wikipedia.org/wiki/Ageing#Theories Ageing](2). Superoxide dismutase is an enzyme that detoxifies superoxide via a special reaction known as dismutation. | ||
Cu/Zn Superoxide dismutase is an important antioxidant defense in nearly all cells exposed to oxygen. | Cu/Zn Superoxide dismutase is an important antioxidant defense in nearly all cells exposed to oxygen. | ||
| Line 9: | Line 10: | ||
==Superoxide Dismutase Family== | ==Superoxide Dismutase Family== | ||
| - | In mammals there are three known isomers of superoxide dismutase (SOD). Copper and Zinc containing SOD1 is located in the cytoplasm. Manganese containing SOD2 is located in the mitochondria while a second Copper and Zinc containing SOD3 is located in the extracellular space. SOD3 and SOD2 are tetramers whereas SOD1 is a dimer. These enzymes perform the dismutation reaction by a similar mechanism. | + | In mammals there are three known isomers of [http://en.wikipedia.org/wiki/Superoxide_dismutase superoxide dismutase] (SOD). Copper and Zinc containing SOD1 is located in the cytoplasm. Manganese containing SOD2 is located in the mitochondria while a second Copper and Zinc containing SOD3 is located in the extracellular space. SOD3 and SOD2 are tetramers whereas SOD1 is a dimer. These enzymes perform the dismutation reaction by a similar mechanism. |
==The Reaction of Dismutase== | ==The Reaction of Dismutase== | ||
| Line 23: | Line 24: | ||
==General Structure== | ==General Structure== | ||
| - | Cu/Zn Superoxide dismutase is a | + | Cu/Zn Superoxide dismutase is a [http://en.wikipedia.org/wiki/Homotetrameric homotetramer], containing four polypeptide chains (B,O,G,Y), 152 residues in length. Each chain contains one alpha-helix and 12-14 beta-sheets. It has a structural weight of 62913.70. It has a ligand identifier of C2 H4 O |
<applet load='2SOD' size='300' frame='true' align='right' caption='Cu/Zn Superoxide Dismutase' /> | <applet load='2SOD' size='300' frame='true' align='right' caption='Cu/Zn Superoxide Dismutase' /> | ||
| Line 45: | Line 46: | ||
RCSB | RCSB | ||
| - | [http://en.wikipedia.org/wiki/ | + | [http://en.wikipedia.org/wiki/Cancer Cancer] |
| - | [http://en.wikipedia.org/wiki/Dismutation dismutation] | ||
| - | [http://en.wikipedia.org/wiki/ | + | |
| + | |||
| + | [http://en.wikipedia.org/wiki/Ageing#Theories Ageing] | ||
Revision as of 20:39, 25 March 2010
Cu/Zn Superoxide Dismutase Cu/Zn Superoxide (SODc) dismutase is an oxidoreductase enzyme, catalyzing the dismutation of superoxide into oxygen and hydrogen peroxide.
Contents |
Introduction
Oxygen is vital to sustain life; our cells cannot live without it. Oxygen is the final acceptor of electrons in respiration at the electron transport chain, allowing us to produce more energy from food. But oxygen is also a dangerous compound (1). Reactive forms of oxygen, such as superoxide, leak from the respiratory chain and wreak havoc on the cell. Superoxide is a free radical; radicals are molecules that readily accept electrons, which make them highly reactive. They can strip electrons from proteins, lipids, or nucleic acids, thereby destroying their functions and resulting in cell dysfunction or death. Free-radical damage has been implicated in Amyotrophic lateral sclerosis (ALS), Arteriosclerosis, Arthritis, Cancer, and Ageing(2). Superoxide dismutase is an enzyme that detoxifies superoxide via a special reaction known as dismutation.
Cu/Zn Superoxide dismutase is an important antioxidant defense in nearly all cells exposed to oxygen. Cu2+ Zn Superoxide Dismutase belongs to the superfamily of oxidoreductases, specifically those acting on superoxide as acceptor.
Superoxide Dismutase Family
In mammals there are three known isomers of superoxide dismutase (SOD). Copper and Zinc containing SOD1 is located in the cytoplasm. Manganese containing SOD2 is located in the mitochondria while a second Copper and Zinc containing SOD3 is located in the extracellular space. SOD3 and SOD2 are tetramers whereas SOD1 is a dimer. These enzymes perform the dismutation reaction by a similar mechanism.
The Reaction of Dismutase
Dismutation is a term that refers to a special type of reaction, where two equal but opposite reactions occur on two separate molecules (1).
The oxidized form of the enzyme is reduced by superoxide to form oxygen. The reduced form of the enzyme, formed in this reaction, then reacts with a second superoxide ion to form peroxide, which takes up two protons along the reaction path to yield hydrogen peroxide.
The hydrogen peroxide formed by superoxide dismutase and by other processes is scavenged by catalase, a ubiquitous heme protein that catalyzes the dismutation of hydrogen peroxide into water and molecular oxygen.
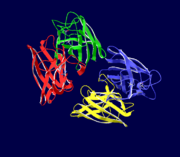
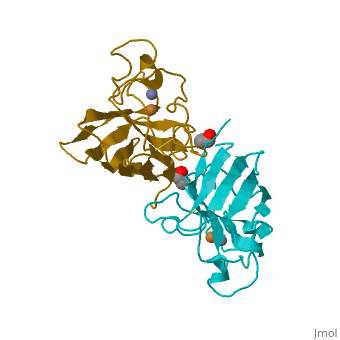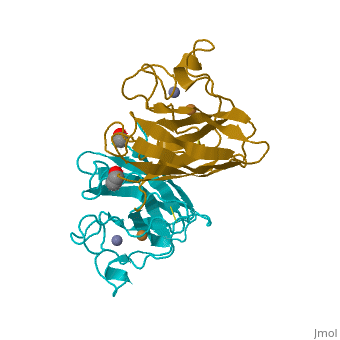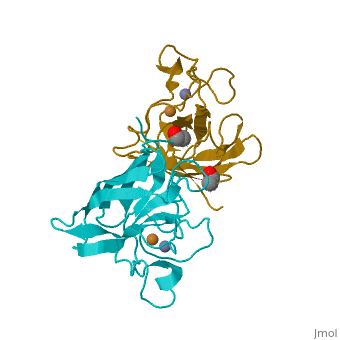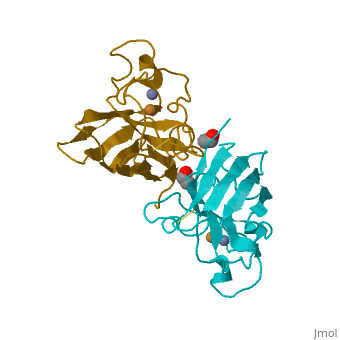
General Structure
Cu/Zn Superoxide dismutase is a homotetramer, containing four polypeptide chains (B,O,G,Y), 152 residues in length. Each chain contains one alpha-helix and 12-14 beta-sheets. It has a structural weight of 62913.70. It has a ligand identifier of C2 H4 O
|
Antioxidant Capabilities
Free radicals such as superoxide are a type of reactive oxygen species that can strip electrons from proteins, lipids, or nucleic acids, thereby destroying their functions and resulting in cell dysfunction or death. Apparently, if SOD is defective, superoxide is not degraded and can destroy cells. Motor neurons appear to be particularly sensitive to superoxide attack.
Importance of Vitamins
Superoxide dismutase and catalase are remarkably efficiently, performing their reactions at or near the diffusion-limited rate. Glutathione peroxide also plays a role in scavenging H2O2. Other cellular defences against oxidative damage include the antioxidant vitamins, vitamins E and C. Because it is lipophilic, vitamin E is especially useful in protecting membranes from lipid peroxidation.
One of the long-term benefits of exercise may be to increase the amount of superoxide dismutase in the cell. The elevated aerobic metabolism during exercise causes more ROS to be generated. In response, the cell synthesizes more protective enzymes. The net effect is one of protection, because the increase in superoxide dismutase more effectively protects the cell during periods of rest.
References
texttext[1]
Seeley, Stephens, Tate. (2008). Anatomy & Physiology 8th Ed.
RCSB
Proteopedia Page Contributors and Editors (what is this?)
Jordan Schibli, Jane S. Richardson, David Canner, Michal Harel, Andrea Gorrell