Ketosteroid Isomerase
From Proteopedia
(→Structural Homologs) |
|||
| Line 34: | Line 34: | ||
===Structural Homologs=== | ===Structural Homologs=== | ||
| - | As noted above, [http://en.wikipedia.org/wiki/Scytalone_dehydratase scytalone dehydratase], [http://en.wikipedia.org/wiki/NUTF2 nuclear transport factor 2 (NTF2)], and [http://en.wikipedia.org/wiki/Naphthalene_1,2-dioxygenase naphthalene 1,2-dioxygenase] share similar structural motifs to ketosteroid isomerase, which facilitate binding of hydrophobic substrates.<ref name="Ha" />,<ref name="Murzin" /> It has also been postulated that bile acid 7α-dehydratase is also a member of this family of protein and that it shares functional similarity with KSI.<ref name="Murzin" /> In a recent paper, Cherney ''et al''<ref name="Cherney">PMID:18589008 </ref>. identified 'Myobacterium tuberculosis' Rv0760c as a structural homolog of KSI. Although these proteins share similar structural motifs there carry out a diverse array of functions.<ref name="Cherney" /> | + | As noted above, [http://en.wikipedia.org/wiki/Scytalone_dehydratase scytalone dehydratase], [http://en.wikipedia.org/wiki/NUTF2 nuclear transport factor 2 (NTF2)], and [http://en.wikipedia.org/wiki/Naphthalene_1,2-dioxygenase naphthalene 1,2-dioxygenase] share similar structural motifs to ketosteroid isomerase, which facilitate binding of hydrophobic substrates.<ref name="Ha" />,<ref name="Murzin" /> It has also been postulated that bile acid 7α-dehydratase is also a member of this family of protein and that it shares functional similarity with KSI.<ref name="Murzin" /> In a recent paper, Cherney ''et al''<ref name="Cherney">PMID:18589008 </ref>. identified ''Myobacterium tuberculosis'' Rv0760c as a structural homolog of KSI. Although these proteins share similar structural motifs there carry out a diverse array of functions.<ref name="Cherney" /> |
==Available Structures== | ==Available Structures== | ||
Revision as of 02:05, 14 April 2010
Contents |
Ketosteroid Isomerase
Introduction
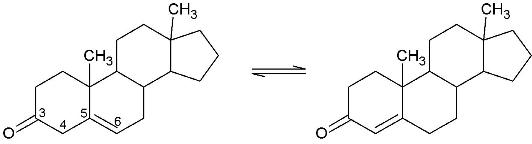
Template:STRUCTURE 1isk (KSI, EC#5.3.3.1) is an enzyme that catalyzes the isomerization of 3-oxo-Δ5 ketosteroids to their hormonally active Δ4-conjugated isomers, as illustrated below.[1], [2]
This reaction is essential in the biosynthesis of steroids in mammals where KSI is a membrane-bound complex.[3] In bacteria, however, KSI exists as a soluble protein is involves in catabolism of steroids.[3] It was first isolated in and has been extensively studied in Commamonas tetosteroni (TI), a bacteria that is capable of growth with testosterone as its sole carbon source.[4] Structural and kinetic studies of this and its homolog from Pseudomonas putida with which it shares 34% sequence and near identical structural homology.[1],[3] It is one of the most efficient known enzymes with an essentially diffusion limited rate of catalysis.[2],[5] It is capable of increasing the catalytic rate by eleven orders of magnitude.[6]
Structure
Ketosteroid isomerase exits as a 28 kDa homodimeric protein, in which the two dimers related to each other via hydrophobic and electrostatic interactions.[5] Each monomer consists of a curved and three . These secondary structures define a conical closed barrel geometry, with one open and one closed end, and create a deep pocket in which the active site resides.[3],[7] This unique geometry is shared by several other proteins (scytalone dehydratase, nuclear transport factor 2, and naphthalene 1,2-dioxygenase), however, these molecules do not share functional or sequence homology. It is speculated that this unique protein structure may enable better binding of hydrophobic substrates such as steroids.[3]
Although the of KSI is notably hydrophobic, it contains several hydrophilic residues believed to be important to the enzymatic function of the protein. The hydrophobic active site of KSI contains an aspartate residue at position 99 and a tyrosine residue at position 14 (according to the numbering for the Commamonas tetosteroni protein, which will be used throughout) that are capable of binding the 3-position carbonyl of the steroid. Additionally, the active site contains an aspartate residue at position 38 that is participates in the catalytic activity of KSI.[1]
Enzyme Mechanism
|
General Mechanism
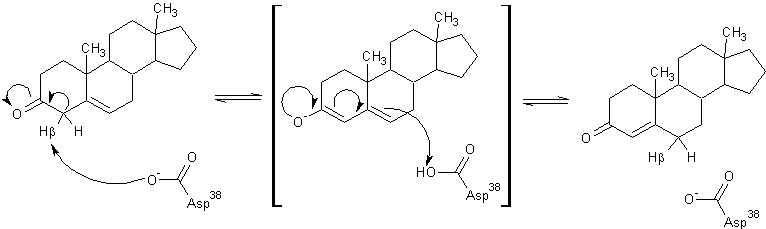
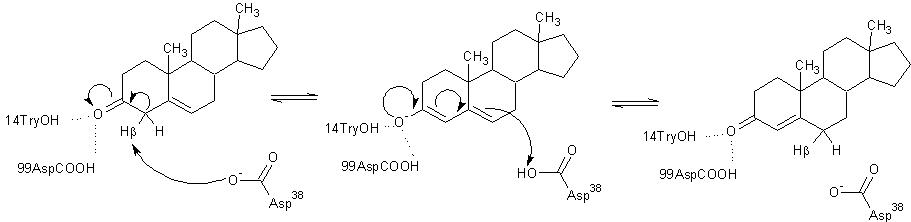
The general mechanism of the proposed reaction of ketosteroid isomerase involves the breaking of a C-H bond adjacent to a carbonyl. This is typically regarded as a difficult reaction due to instability of the intermediate; however it is observed in a number of enzyme-mediated biological reactions.[1] In line with other biological reactions, the mechanism of KSI involves the abstraction the β-hyrdogen from the 4-position carbon resulting in the formation of an enol intermediate, which is followed by reketonization.[1],[3] Structural and kinetic studies suggest that Asp38 (numbering is that of the TI varient of KSI) serves as a general base in this reaction as shown below and abstracts the %beta;-proton from C4 with the syn orbitals of its carboxylate group.[5] Note the formation of the unstable enolate intermediate.
Tyr14 and Asp99 are believed to participate in hydrogen bonding to the O-3 carbonyl of the substrate steroid and stabilize reaction intermediates. Tyr14 is also believed to participate in a low barrier hydrogen bond with the 3-postition oxygen of the steroid, thereby facilitating the abstraction of the β-hydrogen at the 4-position. There are two proposed models of this hydrogen bonding. In , Tyr14 and Asp99 are both bound to the 3-position oxygen, whereas, in , they form a hydrogen bonding network. These are as a single monomer in complex with the intermediate analog equilenin.[8]
Model 1 has become the generally accepted scheme through both crystallographic and mutanagenic/kinetic approaches. Mutation of Asp99 to alanine and Tyr14 to phenylalanine resulted in deleterious effects on kinetic parameters, which were additive in nature suggesting that Tyr14 and Asp99 participate equally in hydrogen binding with the oxygen atom. Additionally, the crystal structure of TI analog from Pseudomonas putida supports this conclusion given the .[1] The general mechanism in the active site of KSI is outlined below.
Low Barrier Hydrogen Bond
Several NMR studies of KSI in complex with substrates have revealed the presence of a highly deshielded proton characteristic of the formation of a low barrier hydrogen bond (LBHB) between Tyr14 and the O-3 atom of various substrates and analogs.[3] The energy of this bond along with that of the normal hydrogen bond from Asp99 most likely contribute to the energy needed to support proton abstraction from the C-4 position.[3]
Related Proteins
Functionally Related Proteins
Ketosteroid isomerase is one of a large number of enzymes that catalyze the cleavage of a C-H bond. Among these are mandelate racemace, triosephosphate isomerase, citrate synthase, and 4-oxalocrotonate tautomerase.[1]
Structural Homologs
As noted above, scytalone dehydratase, nuclear transport factor 2 (NTF2), and naphthalene 1,2-dioxygenase share similar structural motifs to ketosteroid isomerase, which facilitate binding of hydrophobic substrates.[3],[6] It has also been postulated that bile acid 7α-dehydratase is also a member of this family of protein and that it shares functional similarity with KSI.[6] In a recent paper, Cherney et al[9]. identified Myobacterium tuberculosis Rv0760c as a structural homolog of KSI. Although these proteins share similar structural motifs there carry out a diverse array of functions.[9]
Available Structures
- 1isk-First solution phase structure NMR structure of KSI from Comamonas testosteroni
- 1opy
- 1buq
- 1qjg-KSI from Pseudomoas testosteroni in complex with equilenin
- 1c7h
- 1e3r
- 1e3v
- 1e97
- 1ea2
- 1k41
- 1gs3
- 1ogx
- 1oh0-KSI from Pseudomonas putida in complex with equilenin
- 1cqs
- 1vzz
- 1w01
- 1w02
- 1oho-KSI Y16F/D40N mutant from Pseudomonas putida in complex with equilenin
- 1w6y
- 1w0o
- 2pzv
- 2inx
- 3cpo
- 3ex9
- 3fzw
- 3ipt
- 3m8c
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 Pollack RM. Enzymatic mechanisms for catalysis of enolization: ketosteroid isomerase. Bioorg Chem. 2004 Oct;32(5):341-53. PMID:15381400 doi:10.1016/j.bioorg.2004.06.005
- ↑ 2.0 2.1 TALALAY P, WANG VS. Enzymic isomerization of delta5-3-ketosteroids. Biochim Biophys Acta. 1955 Oct;18(2):300-1. PMID:13276386
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 3.7 3.8 Ha NC, Choi G, Choi KY, Oh BH. Structure and enzymology of Delta5-3-ketosteroid isomerase. Curr Opin Struct Biol. 2001 Dec;11(6):674-8. PMID:11751047
- ↑ Ha NC, Choi G, Choi KY, Oh BH. Structure and enzymology of Delta5-3-ketosteroid isomerase. Curr Opin Struct Biol. 2001 Dec;11(6):674-8. PMID:11751047
- ↑ 5.0 5.1 5.2 Wu ZR, Ebrahimian S, Zawrotny ME, Thornburg LD, Perez-Alvarado GC, Brothers P, Pollack RM, Summers MF. Solution structure of 3-oxo-delta5-steroid isomerase. Science. 1997 Apr 18;276(5311):415-8. PMID:9103200
- ↑ 6.0 6.1 6.2 Murzin AG. How far divergent evolution goes in proteins. Curr Opin Struct Biol. 1998 Jun;8(3):380-7. PMID:9666335
- ↑ Cho HS, Choi G, Choi KY, Oh BH. Crystal structure and enzyme mechanism of Delta 5-3-ketosteroid isomerase from Pseudomonas testosteroni. Biochemistry. 1998 Jun 9;37(23):8325-30. PMID:9622484 doi:10.1021/bi9801614
- ↑ Cho HS, Ha NC, Choi G, Kim HJ, Lee D, Oh KS, Kim KS, Lee W, Choi KY, Oh BH. Crystal structure of delta(5)-3-ketosteroid isomerase from Pseudomonas testosteroni in complex with equilenin settles the correct hydrogen bonding scheme for transition state stabilization. J Biol Chem. 1999 Nov 12;274(46):32863-8. PMID:10551849
- ↑ 9.0 9.1 Cherney MM, Garen CR, James MN. Crystal structure of Mycobacterium tuberculosis Rv0760c at 1.50 A resolution, a structural homolog of Delta(5)-3-ketosteroid isomerase. Biochim Biophys Acta. 2008 Nov;1784(11):1625-32. Epub 2008 Jun 6. PMID:18589008 doi:10.1016/j.bbapap.2008.05.012
Proteopedia Page Contributors and Editors (what is this?)
Laura M. Haynes, Michal Harel, Joel L. Sussman, Alexander Berchansky