User:Lynmarie K Thompson/Sandbox 1
From Proteopedia
| Line 8: | Line 8: | ||
Many bacteria can direct their swimming towards favorable environments. They detect molecules such as amino acids or sugars using receptors that bind these molecules and transmit a signal into the cell. This signal controls several proteins which ultimately control the motors that rotate the flagella to cause the cell to either continue swimming or to tumble. When an attractant molecule binds, it signals: "Things look good, keep swimming!" The opposite signal occurs when bacteria sense decreasing concentrations of attractant molecules: "Time to tumble and try a new swimming direction." | Many bacteria can direct their swimming towards favorable environments. They detect molecules such as amino acids or sugars using receptors that bind these molecules and transmit a signal into the cell. This signal controls several proteins which ultimately control the motors that rotate the flagella to cause the cell to either continue swimming or to tumble. When an attractant molecule binds, it signals: "Things look good, keep swimming!" The opposite signal occurs when bacteria sense decreasing concentrations of attractant molecules: "Time to tumble and try a new swimming direction." | ||
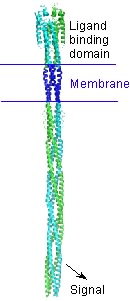
| - | An intact bacterial chemotaxis receptor (see model below) is an unusually long alpha-helical structure. The attractant molecule binds near the top of the receptor and sends a signal across the membrane to control proteins that bind near the bottom of the receptor. | + | An intact bacterial chemotaxis receptor (see model below) is an unusually long alpha-helical structure. The attractant molecule binds near the top of the receptor and sends a signal across the membrane into the cell to control proteins that bind near the bottom of the receptor as pictured. |
[[Image:intactModelLargeText.jpg]] | [[Image:intactModelLargeText.jpg]] | ||
| - | |||
| - | Molecular Playground banner: A receptor protein used by bacteria to sense molecules in their environment. | ||
<applet load='1wat' size='[450,338]' frame='true' align='right' | <applet load='1wat' size='[450,338]' frame='true' align='right' | ||
| Line 18: | Line 16: | ||
The spinning protein (right) is the ligand binding domain of the aspartate receptor protein with aspartate bound. The complete protein (below) is much longer: it extends its alpha helices (from the bottom of the <scene name='User:Lynmarie_K_Thompson/Sandbox_1/Loadedfrompdb/4'>Initial view</scene>) across the membrane into the cell and binds other proteins important for signaling (LKT). | The spinning protein (right) is the ligand binding domain of the aspartate receptor protein with aspartate bound. The complete protein (below) is much longer: it extends its alpha helices (from the bottom of the <scene name='User:Lynmarie_K_Thompson/Sandbox_1/Loadedfrompdb/4'>Initial view</scene>) across the membrane into the cell and binds other proteins important for signaling (LKT). | ||
| + | |||
| + | Molecular Playground banner: A receptor protein used by bacteria to sense molecules in their environment. | ||
Revision as of 21:40, 14 April 2010
CBI Molecules
These are molecules under study by members of the University of Massachusetts Amherst Chemistry-Biology Interface Program. Many of the molecules we study are featured at the Molecular Playground. Follow the links below to read nontechnical descriptions in Proteopedia.
Bacterial chemotaxis receptors, Thompson & Weis laboratories
Many bacteria can direct their swimming towards favorable environments. They detect molecules such as amino acids or sugars using receptors that bind these molecules and transmit a signal into the cell. This signal controls several proteins which ultimately control the motors that rotate the flagella to cause the cell to either continue swimming or to tumble. When an attractant molecule binds, it signals: "Things look good, keep swimming!" The opposite signal occurs when bacteria sense decreasing concentrations of attractant molecules: "Time to tumble and try a new swimming direction."
An intact bacterial chemotaxis receptor (see model below) is an unusually long alpha-helical structure. The attractant molecule binds near the top of the receptor and sends a signal across the membrane into the cell to control proteins that bind near the bottom of the receptor as pictured.
|
The spinning protein (right) is the ligand binding domain of the aspartate receptor protein with aspartate bound. The complete protein (below) is much longer: it extends its alpha helices (from the bottom of the ) across the membrane into the cell and binds other proteins important for signaling (LKT).
Molecular Playground banner: A receptor protein used by bacteria to sense molecules in their environment.