2-Oxoglutarate Dehydrogenase
From Proteopedia
| Line 2: | Line 2: | ||
{{STRUCTURE_ 2jgd | PDB= 2jgd | SCENE= }} | {{STRUCTURE_ 2jgd | PDB= 2jgd | SCENE= }} | ||
| - | + | ==Introduction== | |
The enzyme 2-Oxoglutarate Dehydrogenase E1o is a subunit the 2-Oxoglutarate Dehydrogenase multi enzyme complex. This subunit is a homo-dimer and one of three enzymes that make up the multi-enzyme complex of 2-Oxoglutarate Dehydrogenase. Each of the <scene name='Lucas_Evans_Sandbox/Monomer/2'>monomers</scene> that make up the homo-dimer have an <scene name='Lucas_Evans_Sandbox/Amp1/1'>adenosine monophosphate</scene> cofactor that facilitates the catalysis. E1o is not categorized in the Structural Classification of Proteins (SCOP); however, the <scene name='Lucas_Evans_Sandbox/Secondary_structure/2'>secondary structure</scene> of one of the dimers shows that this enzyme has large sections of alpha-helices followed by a large sections of parallel beta-pleated sheets these alpha and beta subunits are fused as a single polypeptide <ref name="one">PMID:17367808 </ref>. | The enzyme 2-Oxoglutarate Dehydrogenase E1o is a subunit the 2-Oxoglutarate Dehydrogenase multi enzyme complex. This subunit is a homo-dimer and one of three enzymes that make up the multi-enzyme complex of 2-Oxoglutarate Dehydrogenase. Each of the <scene name='Lucas_Evans_Sandbox/Monomer/2'>monomers</scene> that make up the homo-dimer have an <scene name='Lucas_Evans_Sandbox/Amp1/1'>adenosine monophosphate</scene> cofactor that facilitates the catalysis. E1o is not categorized in the Structural Classification of Proteins (SCOP); however, the <scene name='Lucas_Evans_Sandbox/Secondary_structure/2'>secondary structure</scene> of one of the dimers shows that this enzyme has large sections of alpha-helices followed by a large sections of parallel beta-pleated sheets these alpha and beta subunits are fused as a single polypeptide <ref name="one">PMID:17367808 </ref>. | ||
| - | + | ==Catalysis== | |
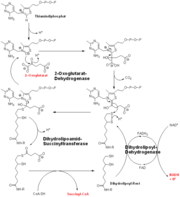
| - | E1o catalyzes the oxidative decarboxylation of alpha-ketoglutarate to Succinyl-CoA at its <scene name='Lucas_Evans_Sandbox/Active_site/1'>active site</scene> in the fourth step of the metabolic citric acid cycle by acting as a base to facilitate the decarboxylation <ref name="one" />. The main residues responsible for the catalysis are thought to be His 260, Phe 227, Gln685, His 729, Ser302, and His 298 <ref name="one" />. E1o is also thought to have a single active site. E1o also requrires two cofactors in order for it to function properly, Thiamine diphosphate and divalent magnesium ion if either are not present then the enzyme has nearly no activity<ref name="two">PMID:192200 </ref>. The specific mexhanism of the E1o subunit are currently unknown; however, There are several theories as to how it functions, among them is the Hexa Uni Ping Pong theory<ref name="two" /> | + | [[Image:oxoglutarate dehydrogenase.png|thumb|left|The complete reaction of the Oxoglutarate Dehydrogenase Multi-Enzyme Complex]]E1o catalyzes the oxidative decarboxylation of alpha-ketoglutarate to Succinyl-CoA at its <scene name='Lucas_Evans_Sandbox/Active_site/1'>active site</scene> in the fourth step of the metabolic citric acid cycle by acting as a base to facilitate the decarboxylation <ref name="one" />. The main residues responsible for the catalysis are thought to be His 260, Phe 227, Gln685, His 729, Ser302, and His 298 <ref name="one" />. E1o is also thought to have a single active site. E1o also requrires two cofactors in order for it to function properly, Thiamine diphosphate and divalent magnesium ion if either are not present then the enzyme has nearly no activity<ref name="two">PMID:192200 </ref>. The specific mexhanism of the E1o subunit are currently unknown; however, There are several theories as to how it functions, among them is the Hexa Uni Ping Pong theory<ref name="two" />. Even though the mechanism isn't fully know the kinetic data have be calculated and are as follows: |
| - | + | * KM: 0.14 ± 0.04 mM | |
| + | * Vmax : 9 ± 3 μmol.min-1.mg-1<ref name="three" >PMID:17657817</ref> | ||
| + | |||
| + | ==Regulation== | ||
E1o catalyzes a rate limiting step in the Kreb's Cycle and lies far form equilibrium (ΔG= -33kJ/mol). As it is a limiting step this makes it a useful reaction to regulate in order to control the Kreb's Cycle. E1o is inhibited by both NADH and Succinyl-CoA via non competitive feedback inhibition<ref name="one" />. | E1o catalyzes a rate limiting step in the Kreb's Cycle and lies far form equilibrium (ΔG= -33kJ/mol). As it is a limiting step this makes it a useful reaction to regulate in order to control the Kreb's Cycle. E1o is inhibited by both NADH and Succinyl-CoA via non competitive feedback inhibition<ref name="one" />. | ||
| - | + | ==Transfer to E2o== | |
The overall complex (all of the sub units) help catalysis by keeping the necessary substrates for the reaction close within the enzyme so that it is more likely that the substrate will be in favorable conformation. This enzyme is also part of a larger multienzyme complex that channels the intermediates in the catalysis between subunits of the complex thus minimizing unwanted side reactions<ref>Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry: Life at the Molecular Level. Hoboken, NJ: Wiley, 2008. p.580</ref>. Not only do the subunits ferry products back and forth but each of the mers in the E1o homodimer are connected via a cavity lined with acidic residues thus increasing the dimer's ability to act as a base. | The overall complex (all of the sub units) help catalysis by keeping the necessary substrates for the reaction close within the enzyme so that it is more likely that the substrate will be in favorable conformation. This enzyme is also part of a larger multienzyme complex that channels the intermediates in the catalysis between subunits of the complex thus minimizing unwanted side reactions<ref>Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry: Life at the Molecular Level. Hoboken, NJ: Wiley, 2008. p.580</ref>. Not only do the subunits ferry products back and forth but each of the mers in the E1o homodimer are connected via a cavity lined with acidic residues thus increasing the dimer's ability to act as a base. | ||
Revision as of 14:33, 23 April 2010
Contents |
2-Oxoglutarate Dehydrogenase E1o
Introduction
The enzyme 2-Oxoglutarate Dehydrogenase E1o is a subunit the 2-Oxoglutarate Dehydrogenase multi enzyme complex. This subunit is a homo-dimer and one of three enzymes that make up the multi-enzyme complex of 2-Oxoglutarate Dehydrogenase. Each of the that make up the homo-dimer have an cofactor that facilitates the catalysis. E1o is not categorized in the Structural Classification of Proteins (SCOP); however, the of one of the dimers shows that this enzyme has large sections of alpha-helices followed by a large sections of parallel beta-pleated sheets these alpha and beta subunits are fused as a single polypeptide [1].
Catalysis
E1o catalyzes the oxidative decarboxylation of alpha-ketoglutarate to Succinyl-CoA at its in the fourth step of the metabolic citric acid cycle by acting as a base to facilitate the decarboxylation [1]. The main residues responsible for the catalysis are thought to be His 260, Phe 227, Gln685, His 729, Ser302, and His 298 [1]. E1o is also thought to have a single active site. E1o also requrires two cofactors in order for it to function properly, Thiamine diphosphate and divalent magnesium ion if either are not present then the enzyme has nearly no activity[2]. The specific mexhanism of the E1o subunit are currently unknown; however, There are several theories as to how it functions, among them is the Hexa Uni Ping Pong theory[2]. Even though the mechanism isn't fully know the kinetic data have be calculated and are as follows:- KM: 0.14 ± 0.04 mM
- Vmax : 9 ± 3 μmol.min-1.mg-1[3]
Regulation
E1o catalyzes a rate limiting step in the Kreb's Cycle and lies far form equilibrium (ΔG= -33kJ/mol). As it is a limiting step this makes it a useful reaction to regulate in order to control the Kreb's Cycle. E1o is inhibited by both NADH and Succinyl-CoA via non competitive feedback inhibition[1].
Transfer to E2o
The overall complex (all of the sub units) help catalysis by keeping the necessary substrates for the reaction close within the enzyme so that it is more likely that the substrate will be in favorable conformation. This enzyme is also part of a larger multienzyme complex that channels the intermediates in the catalysis between subunits of the complex thus minimizing unwanted side reactions[4]. Not only do the subunits ferry products back and forth but each of the mers in the E1o homodimer are connected via a cavity lined with acidic residues thus increasing the dimer's ability to act as a base.
References
- ↑ 1.0 1.1 1.2 1.3 Frank RA, Price AJ, Northrop FD, Perham RN, Luisi BF. Crystal structure of the E1 component of the Escherichia coli 2-oxoglutarate dehydrogenase multienzyme complex. J Mol Biol. 2007 May 4;368(3):639-51. Epub 2007 Feb 7. PMID:17367808 doi:10.1016/j.jmb.2007.01.080
- ↑ 2.0 2.1 McMinn CL, Ottaway JH. Studies on the mechanism and kinetics of the 2-oxoglutarate dehydrogenase system from pig heart. Biochem J. 1977 Mar 1;161(3):569-81. PMID:192200
- ↑ Leung PS, Rossaro L, Davis PA, Park O, Tanaka A, Kikuchi K, Miyakawa H, Norman GL, Lee W, Gershwin ME. Antimitochondrial antibodies in acute liver failure: implications for primary biliary cirrhosis. Hepatology. 2007 Nov;46(5):1436-42. PMID:17657817 doi:10.1002/hep.21828
- ↑ Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry: Life at the Molecular Level. Hoboken, NJ: Wiley, 2008. p.580
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, David Canner, Lucas Evans, Alexander Berchansky, Joel L. Sussman