Sandbox Prolyl Hydroxylase Domain (PHD) Enzyme
From Proteopedia
(New page: One of the CBI Molecules being studied in the [http://www.umass.edu/cbi/ University of Massachusetts Amherst Chemistry-Biology Interface Program] at UMass Amherst and on display at th...) |
|||
| Line 1: | Line 1: | ||
One of the [[CBI Molecules]] being studied in the [http://www.umass.edu/cbi/ University of Massachusetts Amherst Chemistry-Biology Interface Program] at UMass Amherst and on display at the [http://www.molecularplayground.org/ Molecular Playground]. | One of the [[CBI Molecules]] being studied in the [http://www.umass.edu/cbi/ University of Massachusetts Amherst Chemistry-Biology Interface Program] at UMass Amherst and on display at the [http://www.molecularplayground.org/ Molecular Playground]. | ||
| - | Metazoans adapt to oxygen levels in the environment by making use of these intracellular oxygen levels as signals to regulate the [http://en.wikipedia.org/wiki/Transcription_(genetics) transcription] of genes essential under normoxic or hypoxic conditions. Central to this mechanism is the oxygen-dependent hydroxylation on specific proline and asparagine residues of hypoxia-inducible factor (HIF)-alpha. | + | Metazoans adapt to oxygen levels in the environment by making use of these intracellular oxygen levels as signals to regulate the [http://en.wikipedia.org/wiki/Transcription_(genetics) transcription] of genes essential under normoxic or [http://en.wikipedia.org/wiki/Hypoxia_(medical) hypoxic] conditions. Central to this mechanism is the oxygen-dependent hydroxylation on specific proline and asparagine residues of hypoxia-inducible factor [http://en.wikipedia.org/wiki/HIF1A (HIF)-alpha]. |
| - | Prolyl hydroxylase domain (PHD) enzyme (EC 1.14.11) is a Fe(II)/2-oxoglutarate-dependent dioxygenase that catalyzes the trans-4-hydroxylation of the specific proline residues (either Pro-402 or Pro-564) in the transcription factor, (HIF)-alpha. | + | Prolyl hydroxylase domain (PHD) enzyme [http://www.chem.qmul.ac.uk/iubmb/enzyme/EC1/14/11/ (EC 1.14.11.-)] is a Fe(II)/2-oxoglutarate-dependent dioxygenase that catalyzes the trans-4-hydroxylation of the specific proline residues (either Pro-402 or Pro-564) in the transcription factor, (HIF)-alpha. |
Revision as of 18:02, 30 April 2010
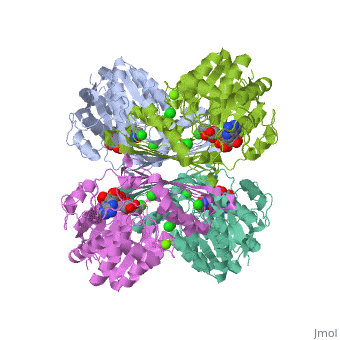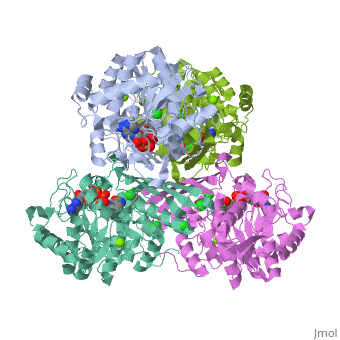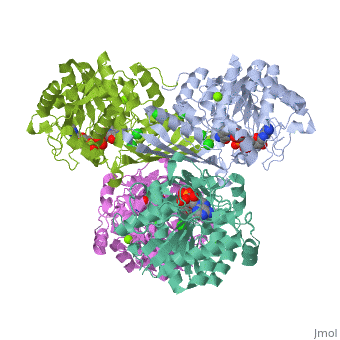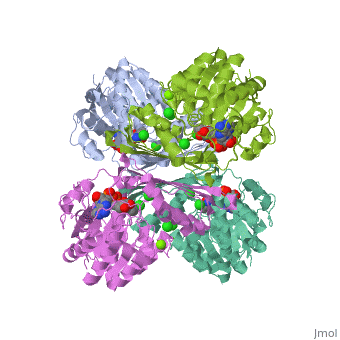
One of the CBI Molecules being studied in the University of Massachusetts Amherst Chemistry-Biology Interface Program at UMass Amherst and on display at the Molecular Playground.
Metazoans adapt to oxygen levels in the environment by making use of these intracellular oxygen levels as signals to regulate the transcription of genes essential under normoxic or hypoxic conditions. Central to this mechanism is the oxygen-dependent hydroxylation on specific proline and asparagine residues of hypoxia-inducible factor (HIF)-alpha.
Prolyl hydroxylase domain (PHD) enzyme (EC 1.14.11.-) is a Fe(II)/2-oxoglutarate-dependent dioxygenase that catalyzes the trans-4-hydroxylation of the specific proline residues (either Pro-402 or Pro-564) in the transcription factor, (HIF)-alpha.
| |||||||||
| 3cin, resolution 1.70Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , , | ||||||||
| Gene: | TM1419, TM_1419 (Thermotoga maritima MSB8) | ||||||||
| Activity: | Inositol-3-phosphate synthase, with EC number 5.5.1.4 | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum, TOPSAN | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
|
Ligand-binding domain
The spinning protein () ) is the ligand binding domain of the aspartate receptor with the aspartate ligand bound (LKT).