Ferredoxin
From Proteopedia
| Line 38: | Line 38: | ||
3dqy, 2qpz - Fd – Pseudomonas putida <br /> | 3dqy, 2qpz - Fd – Pseudomonas putida <br /> | ||
| - | 4Fe-4S containing ferredoxins | + | ===4Fe-4S containing ferredoxins === |
3eun – AvFd – Allochromatium vinosum <br /> | 3eun – AvFd – Allochromatium vinosum <br /> | ||
| Line 51: | Line 51: | ||
1vjw – Fd – Thermotoga maritima <br /> | 1vjw – Fd – Thermotoga maritima <br /> | ||
| - | 3Fe-4S containing ferredoxins | + | ===3Fe-4S containing ferredoxins=== |
2v2k – Fd – Mycobacterium smegmatis <br /> | 2v2k – Fd – Mycobacterium smegmatis <br /> | ||
Revision as of 06:56, 27 May 2010
|
3D structure of acetylcholinesterase
Ferredoxin
Ferredoxin (Fd) is found in chloroplasts which mediates electron transfer and contains an iron-sulfur cluster. It is involved in the photosynthesis process where its iron atoms accept or discharge electrons when they are being oxidized or reduced. The iron-sulfur cluster can contain 2Fe-2S and is termed plant-like or 3Fe-4S or 4Fe-4S clusters. Adrenodoxin (ADR) is a ferredoxin containing a 2Fe-2S group involved in electron transfer from NADPH+ to a cytochrome P-450 in the adrenal gland. Putidaredoxin (PUT) and terpredoxin (TER) are involved in the same reaction in bacteria and contain a 2Fe-2S group.
2Fe-2S containing ferredoxins
3hui – Fd – Rhodopseudomonas palustris
2kaj, 1dox, 1doy – SyFd +Ga – Synechocystis – NMR
1off – SyFd
3gce – Fd – Nocardioides aromaticivorans
2e4p, 2e4q - Fd – Pseudomonas sp.
2q3w, 1vm9 - PmFd (mutant) – Pseudomonas mendocina
2i7f - Fd – Rhodobacter capsulatus
1rfk - Fd – Cyanobacterium masticogladus laminosus
1vck - Fd – Pseudomonas resinovorans
1wri, 1frr - Fd – Equisetum arvense
1sjg - PmFd– NMR
1iue - Fd– Plasmodium falciparum
1m2a –AeFd – Aquifex aeolicus
1m2b, 1m2d, 1f37, 1f5b, 1f5c – AeFd (mutant)
1l5p – Fd – Trichomonas vaginalis
1i7h - Fd – Escherichia coli
1czp, 1qt9, 1frd, 1fxa - aFd– anabaena
1j7a, 1j7b, 1j7c , 1qoa, 1qob, 1qof, 1qog- aFd (mutant)
1e0z – Fd – Halobacterium salinarium
1pfd – Fd – Petroselinum crispum – NMR
1a70 - Fd (mutant) – Spinacia oleracea
1awd - Fd – Chlorella fusca
2cjn, 2cjo, 1roe – SyFd – NMR
1rof – SyFd – Synechococcus elongates
1doi - Fd – Haloarcula marismortui
4fxc – Fd – Spirulina platensis
1fxi – Fd – Aphanothece sacrum
3dqy, 2qpz - Fd – Pseudomonas putida
4Fe-4S containing ferredoxins
3eun – AvFd – Allochromatium vinosum
3exy - AvFd (mutant)
2vkr - Fd+Zn – Acidianus ambivalens
2z8q - PfFd (mutant) – Pyrococcus furiosus
2fgo - Fd– Pseudomonas aeruginosa
1iqz, 1ir0 - BtFd – Bacillus thermoproteolyticus
1rgv - Fd – Thauera aromatica
1dax, 1dfd – DaFd – Desulfovibrio africanus – NMR
1fxr - DaFd
1vjw – Fd – Thermotoga maritima
3Fe-4S containing ferredoxins
2v2k – Fd – Mycobacterium smegmatis
1wtf - BtFd (mutant)
1sj1 - PfFd
1fxd - DgFd – Desulfovibrio gigas
1f2g – DgFd – NMR
1xer - Fd – Sulfolobus tokodaii
4Fe-4S+3Fe-4S containing ferredoxins
1gao, 6fdr, 7fd1, 7fdr, 1axq, 6fd1, 1frh, 1fri, 1frj, 1frk, 1frl, 1frm, 1fda, 1fdb, 1fdd, 5fd1, 1fer – AvFd – Azotobacter vinelandii
1pc4, 1pc5, 1g6b, 1g3o, 1ff2, 1b0v, 1d3w, 1b0t, 1a6l, 1ftc, 1frx, 2fd2, 1fd2 - AvFd (mutant)
1h98 – Fd – Thermus thermophilus
1a8p, 1bd6 - BsFd – Bacillus schlegelii – NMR
4Fe-4S+4Fe-4S containing ferredoxins
1dur – Fd – Peptoniphilus asaccarolyticus
1bwe, 1bqx - BsFd (mutant) – NMR
2fdn, 1fca , 1fdn- Fd – Clostridium acidi-urici
1blu - Fd – Chromatium vinosum
1clf – Fd – Clostridium pasteurianum – NMR
Adrenoredoxin
2jqr – ADR Fd domain (mutant)+cytochrome c (mutant) – yeast – NMR
2bt6 – cADR1 modified – cow
1l6u, 1l6v – cADR1 – NMR
1e6e – cADR (mutant)+ADR reductase
1cje, 1ayf - cADR
Putidaredoxin
1yji, 1yjj, 1pdx – PpPUT – Pseudomonas putida – NMR
3lb8 – PpPUT (mutant)+PUT reductase
1xln, 1xlo, 1xlp, 1xlq, 1r7s, 1oqq, 1oqr - PpPUT (mutant)
1gpx, 1put- PpPUT (mutant) - NMR
terpredoxin
1b9r – TER – Pseudomonas - NMR
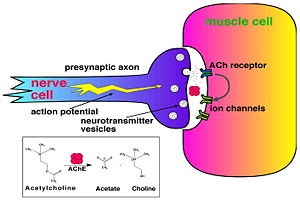
Acetylcholinesterase (AChE) is key enzyme in the nervous system of animals. By rapid hydrolysis of the neurotransmitter, acetylcholine (ACh), AChE terminates neurotransmission at cholinergic synapses. It is a very fast enzyme, especially for a serine hydrolase, functioning at a rate approaching that of a diffusion-controlled reaction. AChE inhibitors are among the key drugs approved by the FDA for management of Alzheimer's disease (AD). The powerful toxicity of organophosphorus (OP) poisons is attributed primarily to their potent AChE inhibitors.
The 3D structure of Torpedo californica AChE (TcAChE) (Sussman et al. & Silman (1991)) opened up new horizons in research on an enzyme that had already been the subject of intensive investigation. The unanticipated structure of this extremely rapid enzyme, in which the active site was found to be buried at the bottom of a , lined by aromatic residues, led to a revision of the views then held concerning substrate traffic, recognition, and hydrolysis (Botti et al. Sussman & Silman (1999)). To understand how those aromatic residues behave with the enzyme, see Flexibility of aromatic residues in acetylcholinesterase.
Alzheimer’s disease (AD) is a debilitating brain disease that occurs in around 10% of the elderly and, as yet, there is no known cure. At present, the most widely used treatments consist are medications that attempt to increase the brain’s levels of ACh, whose levels decrease with onset of disease. These drugs work by interfering with AChE. Thus drugs that are mild inhibitors of AChE, like Tacrine, E2020 (Aricept) and the Traditonal Chinese Medicine (TCM) Huperzine appear to retard symptoms of AD.
|
The active site gorge has , a catalytic site (consisting of the catalytic triad together with Trp84 & Phe330) and a peripheral site (including Trp 279 & Tyr 121), which helps prebind the substrate and direct it toward the active site. The 3D structure showed not only that the active site was buried deep in the enzyme, but surprisingly, there were no negatively charged residues along this gorge, as was expected to help attract the positively charged ACh substrate, rather, instead, a series of aromatic residues that are highly conserved in all AChE sequences. See: AChE inhibitors and substrates
Selected 3D Structures of AChE
Acetylcholinesterase - AChE native
3lii – hAChE - recombinant human
1ea5, 2ace – TcAChE – trigonal – Torpedo californica
2j3d – TcAChE – monoclinic
1w75 – TcAChE – orthorhombic
1eea – TcAChE – cubic
2vt6, 2vt7 – TcAChE – different dosage
1qid to 1qim - TcAChE synchrotron radiation damage
1j06, 1maa – mAChE - mouse
1qo9 – DmAChE - Drosophila
1c2o, 1c2b – electrophorus AChE – Electric eel
AChE active site inhibitors conjugating at the bottom of the active site gorge
2w9i – TcAChE + methylene blue
2wls – MosAChE + AMTS13
2vq6 – TcAChE + 2-PAM
2j3q – TcAChE + Thioflavin T
2ha0 – mAChE + ketoamyltrimethylammonium
2h9y – mAChE + TMTFA
1gpk, 1gpn, 1vot – TcAChE + huperzine
1gqr – TcAChE + rivastigmine
1gqs – TcAChE + NAP
1e66 – TcAChE + huprine
1dx4, 1qon – DmAChE + tacrine derivative
1oce – TcAChE + MF268
1ax9, 1ack – TcAChE + edrophonium
1amn – TcAChE + TMTFA
1acj – TcAChE + tacrine
AChE peripheral site inhibitors conjugating at the surface of the protein
1ku6 - mAChE + fasciculin 2
1ku6, 1mah - mAChE + fasciculin 2
1j07 - mAChE + decidium
1n5m - mAChE + gallamine
1n5r - mAChE + propidium
1b41, 1f8u - hAChE + fasciculin 2
1fss - TcAChE + fasciculin 2
AChE bis inhibitors spanning the active site gorge
3i6m – TcAChE + N-piperidinopropyl galanthamine
3i6z - TcAChE + saccharinohexyl galanthamine
1zgb, 1zgc – TcAChE + tacrine (10) hupyridone
2w6c – TcAChE + bis-(-)-nor-meptazinol
2ckm, 2cmf – TcAChE + bis-tacrine
2cek – TcAChE + N-[8-(1,2,3,4-tetrahydroacridin-9-ylthio)octyl]-1,2,3,4-tetrahydroacridin-9-amine
1ut6 - TcAChE + N-9-(1,2,3,4-tetrahydroacridinyl)-1,8-diaminooctane
1odc - TcAChE + N-4-quinolyl-N-9-(1,2,3,4-tetrahydroacridinyl)-1,8-diaminooctane
1w4l, 1w6r, 1w76, 1dx6, 1qti - TcAChE + galanthamine and derivative
1q83, 1q84 - mAChE + TZ2PA6
1h22, 1h23 – TcAChE + bis-hupyridone
1hbj – TcAChE + quinoline derivativev
1e3q – TcAChE + bw284c51
1eve – TcAChE + e2020
1acl – TcAChE + decamethonium
AChE organophosphate inhibitors causing irreversible inhibition
2wu3 – mAChE + fenamiphos and HI-6
2wu4 – mAChE + fenamiphos and ortho-7
2jgf - mAChE + fenamiphos
2wfz, 2wg0, 1som - TcAChE + soman
2wg1 - TcAChE + soman + 2-PAM
2whp, 2whq, 2whr – mAChE + sarin and HI-6
2jgg - mAChE + sarin
2jgl - mAChE + VX and sarin
1cfj - TcAChE + sarin, GB
3dl4, 3dl7 – mAChE + tabun
2jey – mAChE + HLO-7
2c0p, 2c0q - mAChE + tabun
2jez - mAChE + tabun + HLO-7
2jf0 - mAChE + tabun + Ortho-7
2jgh - mAChE + VX
1vxo, 1vxr - TcAChE + VX
2jgi, 2jgm - mAChE + DFP
1dfp - TcAChE + DFP
2jgj, 2jgk, 2jge - mAChE + methamidophos
2gyu - mAChE + HI-6
2gyv - mAChE + Ortho-7
2gyw - mAChE + obidoxime
AChE substrate analogues mimicking the binding of the substrate acetylcholine
2ha4 – mAChE (mutant) + acetylcholine
2vja, 2vjb, 2vjc, 2vjd, 2cf5 – TcAChE + 4-oxo-N,N,N-trimethylpentanaminium
2v96, 2v97, 2v98, 2v99 – TcAChE + 1-(2-nitrophenyl)-2,2,2-trifluoroethyl-arsenocholine
2ha2 – mAChE + succinylcholine
2ha3 - mAChE + choline
2ha5 – mAChE (mutant) + acetylthiocholine
2ha6 – mAChE (mutant) + succinylthiocholine
2ha7 – mAChE (mutant) + butyrylthiocholine
2ch4, 2c58 – TcAChE + acetylthiocholine
2c5g – TcAChE + thiocholine
Others...
2j4f – TcAChE + Hg
1vzj – TcAChE tetramerization domain
1jjb – TcAChE + PEG
More structures can be obtained by searching for
AChE
External Links
Acetylcholinesterase Tutorial by Karl Oberholser, Messiah College
PDB Molecule of the Month - Acetylcholinesterase
Movies: X-ray Damage in ACh & Nature's Vacuum Cleaner by R. Gillilan, Cornell Univ
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Joel L. Sussman, Jaime Prilusky, Eran Hodis, Wayne Decatur, David Canner