This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Sandbox 16
From Proteopedia
| Line 6: | Line 6: | ||
== Structure == | == Structure == | ||
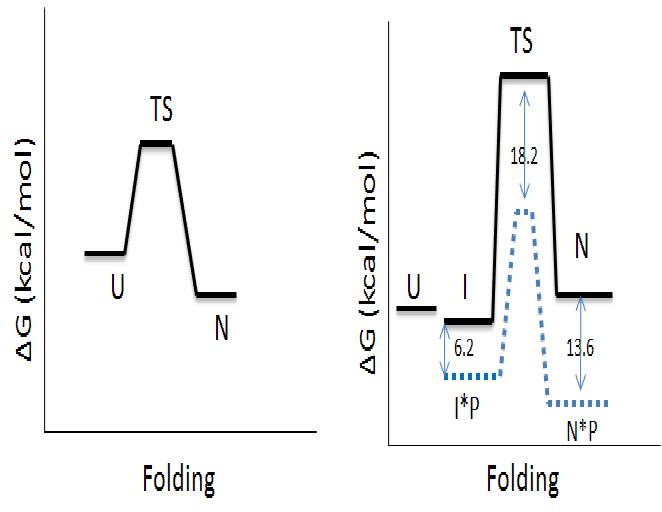
| - | <p>The <scene name='Sandbox_16/Alp-1/2'>secondary structure</scene> contains the double β-barrel motif (colored yellow) that is characteristic of the chymotrypsin family as well as an active site containing the <scene name='Sandbox_16/Alp-1/6'>"catalytic triad"</scene> - His 57, Asp 102, and Ser 195 - that is responsible for proteolysis. The preference for αLP to cleave substrates on the C-terminal side of small hydrophobic residues, such as Alanine and Valine is mostly due to <scene name='Sandbox_16/Alp-1/3'>three residues in the S1 pocket</scene> consisting of Met 190, Met 213, and Val 218<ref>PMID:9232638</ref>. In contrast to its mammalian homologs like trypsin and chymotrypsin, αLP is synthesized with a 166 residue N-terminal Pro region that plays an obligatory role in the proper folding of its 198 residue protease domain<ref>PMID:2646278</ref>. The barrier to folding | + | <p>The <scene name='Sandbox_16/Alp-1/2'>secondary structure</scene> contains the double β-barrel motif (colored yellow) that is characteristic of the chymotrypsin family as well as an active site containing the <scene name='Sandbox_16/Alp-1/6'>"catalytic triad"</scene> - His 57, Asp 102, and Ser 195 - that is responsible for proteolysis. The preference for αLP to cleave substrates on the C-terminal side of small hydrophobic residues, such as Alanine and Valine is mostly due to <scene name='Sandbox_16/Alp-1/3'>three residues in the S1 pocket</scene> consisting of Met 190, Met 213, and Val 218<ref>PMID:9232638</ref>. In contrast to its mammalian homologs like trypsin and chymotrypsin, αLP is synthesized with a 166 residue N-terminal Pro region that plays an obligatory role in the proper folding of its 198 residue protease domain<ref>PMID:2646278</ref>. The Pro region overcomes the barrier to folding by providing a catalyzed pathway in which the transition state to folding is lowered by 18.2 kcal/mol<ref>PMID:9796818</ref>. The product of this folding is not active αLP but an inhibitory complex, N*P. The release of active αLP requires the removal of the Pro region via proteolysis, which occurs naturally. This leaves the native αLP, a metastable state with a large barrier to unfolding (t<sub>1/2</sub>~1.2 years). Below shows the free energy diagram summarizing the difference between the folding landscape of a typical thermodynamically stable protein (left) and that of αLP (right). The dotted blue line indicates the folding landscape in the presence of the Pro region. |
[[Image:therm vs kinetic.jpg]] | [[Image:therm vs kinetic.jpg]] | ||
Revision as of 16:48, 23 June 2010
Alpha-lytic protease
Alpha-lytic protease (αLP) is a 198 residue extracellular bacterial serine protease produced by Lysobacter enzymogenes. The three-dimensional fold of αlp puts it in the same class as cymotrypsin, trypsin and other digestive serine proteases despite only modest sequence homology[1]. However, unlike its thermodynamically stable homologs, αLP is stabilized by a large unfolding activation barrier. This kinetic stability optimizes the native state to survive under the harsh, proteolytic conditions in which it operates. Since the native state is less stable than both an intermediate and a completely unfolded state, αLP requires a Pro region to facilitate folding by stabilizing the folding transition state as well as the native state. After folding, the pro region is proteolytically cleaved, leaving an active αLP kinetically trapped
| |||||||||
| 2alp, resolution 1.70Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | |||||||||
| Activity: | Alpha-lytic endopeptidase, with EC number 3.4.21.12 | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Structure
The contains the double β-barrel motif (colored yellow) that is characteristic of the chymotrypsin family as well as an active site containing the - His 57, Asp 102, and Ser 195 - that is responsible for proteolysis. The preference for αLP to cleave substrates on the C-terminal side of small hydrophobic residues, such as Alanine and Valine is mostly due to consisting of Met 190, Met 213, and Val 218[2]. In contrast to its mammalian homologs like trypsin and chymotrypsin, αLP is synthesized with a 166 residue N-terminal Pro region that plays an obligatory role in the proper folding of its 198 residue protease domain[3]. The Pro region overcomes the barrier to folding by providing a catalyzed pathway in which the transition state to folding is lowered by 18.2 kcal/mol[4]. The product of this folding is not active αLP but an inhibitory complex, N*P. The release of active αLP requires the removal of the Pro region via proteolysis, which occurs naturally. This leaves the native αLP, a metastable state with a large barrier to unfolding (t1/2~1.2 years). Below shows the free energy diagram summarizing the difference between the folding landscape of a typical thermodynamically stable protein (left) and that of αLP (right). The dotted blue line indicates the folding landscape in the presence of the Pro region.
References
- ↑ Brayer GD, Delbaere LT, James MN. Molecular structure of the alpha-lytic protease from Myxobacter 495 at 2.8 Angstroms resolution. J Mol Biol. 1979 Jul 15;131(4):743-75. PMID:117110
- ↑ Rader SD, Agard DA. Conformational substates in enzyme mechanism: the 120 K structure of alpha-lytic protease at 1.5 A resolution. Protein Sci. 1997 Jul;6(7):1375-86. PMID:9232638
- ↑ Silen JL, Frank D, Fujishige A, Bone R, Agard DA. Analysis of prepro-alpha-lytic protease expression in Escherichia coli reveals that the pro region is required for activity. J Bacteriol. 1989 Mar;171(3):1320-5. PMID:2646278
- ↑ Sohl JL, Jaswal SS, Agard DA. Unfolded conformations of alpha-lytic protease are more stable than its native state. Nature. 1998 Oct 22;395(6704):817-9. PMID:9796818 doi:10.1038/27470

