User:Wayne Decatur/Sandbox Glutamate receptor
From Proteopedia
m (→About this Structure) |
m (added static image colored like in journal article on 3kg2) |
||
| Line 1: | Line 1: | ||
| - | [[Image:3KG2-snapshot-900x900- | + | [[Image:3KG2-snapshot-900x900-14724.jpg|left|300px]] |
<!-- | <!-- | ||
Revision as of 01:50, 2 July 2010
Contents |
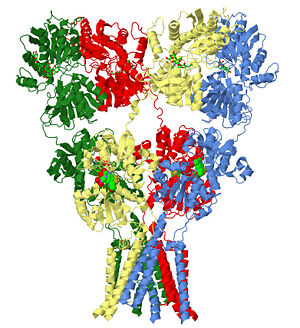
AMPA subtype ionotropic glutamate receptor in complex with competitive antagonist ZK 200775
Template:ABSTRACT PUBMED 19946266
About this Structure
| |||||||
| glutamate receptor (3kg2), resolution 3.6Å () | |||||||
|---|---|---|---|---|---|---|---|
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
3kg2 is a 4 chains structure of sequences from Rattus norvegicus. Full crystallographic information is available from OCA.
REDO IMAGES SO LOOK CLOSER TO COLORS IN ACCOMPANYING ARTICLE. (OR AT LEAST MAKE SO DEFAULT JMOL COLORS MATCH ORDER USED ON PAGE SINCE THEY AREN'T TOO FAR OFF.) MEANS I NEED TO REDO THE JENA PAGE AT THE TOP TOO. but I can leave the one I first made for the 3kg2 on that page so first figure made is usable on another page.
.
To help give a better idea of how the glutamate receptor is oriented on the cell surface in the membrane lipid bilayer, as calculated by the Orientations of Proteins in Membranes database(University of Michigan, USA) is shown with the red atoms indicating the boundary of the hydrophobic core closet to the outside of the cell and the blue atoms indicating the boundary closest to the inside of the cell.
Related Structures
- 3kgc GluA2 ligand-binding core complex bound with glutamate
- 2a5t GluN1-GluN2A ligand-binding domain heterodimer
- 2a5s GluN2A ligand-binding domain bound with glutamate
See Also
Page started with original page seeded by OCA on Wed Dec 16 11:24:54 2009 for 3kg2.