User:Wayne Decatur/Sandbox Glutamate receptor
From Proteopedia
m |
m |
||
| Line 17: | Line 17: | ||
--> | --> | ||
{{ABSTRACT_PUBMED_19946266}} | {{ABSTRACT_PUBMED_19946266}} | ||
| + | ==Background== | ||
| + | Citations <ref>PMID:19946266</ref><ref>PMID: 20010675</ref> . | ||
==About this Structure== | ==About this Structure== | ||
| Line 28: | Line 30: | ||
|RELATEDENTRY= | |RELATEDENTRY= | ||
|RESOURCES=<span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=3kg2 FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=3kg2 OCA], [http://www.ebi.ac.uk/pdbsum/3kg2 PDBsum], [http://www.rcsb.org/pdb/explore.do?structureId=3kg2 RCSB]</span> | |RESOURCES=<span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=3kg2 FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=3kg2 OCA], [http://www.ebi.ac.uk/pdbsum/3kg2 PDBsum], [http://www.rcsb.org/pdb/explore.do?structureId=3kg2 RCSB]</span> | ||
| - | }} | + | }}The homomeric rat GluA2 receptor <scene name='User:Wayne_Decatur/Sandbox_Glutamate_receptor/Default3kg2/1'>has four subunits that are arranged to form a 'Y'-shape</scene> with the <scene name='User:Wayne_Decatur/Sandbox_Glutamate_receptor/Meas3kg2/1'>'top' being about three times the width of the 'bottom'</scene><ref>PMID: 19946266</ref>. |
| - | The homomeric rat GluA2 receptor <scene name='User:Wayne_Decatur/Sandbox_Glutamate_receptor/Default3kg2/1'>has four subunits that are arranged to form a 'Y'-shape</scene> with the <scene name='User:Wayne_Decatur/Sandbox_Glutamate_receptor/Meas3kg2/1'>'top' being about three times the width of the 'bottom'</scene><ref>PMID: 19946266</ref>. | + | |
====Domains==== | ====Domains==== | ||
| - | The subunits themselves are modular <ref>PMID: 7539962</ref> and in the major domains are found in layers in the tetrameric structure. | + | The subunits themselves are modular <ref>PMID: 7539962</ref>and in the major domains are found in layers in the tetrameric structure. |
::The 'top' layer is composed of the | ::The 'top' layer is composed of the | ||
| Line 59: | Line 60: | ||
==References== | ==References== | ||
| - | + | {{Reflist}} | |
''Page started with original page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Wed Dec 16 11:24:54 2009 for [[3kg2]].'' | ''Page started with original page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Wed Dec 16 11:24:54 2009 for [[3kg2]].'' | ||
Revision as of 01:42, 4 July 2010
Contents |
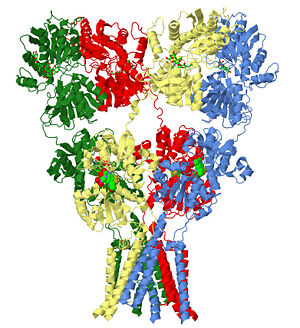
AMPA subtype ionotropic glutamate receptor in complex with competitive antagonist ZK 200775
Template:ABSTRACT PUBMED 19946266
Background
About this Structure
| |||||||
| glutamate receptor (3kg2), resolution 3.6Å () | |||||||
|---|---|---|---|---|---|---|---|
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
The homomeric rat GluA2 receptor with the [3].
Domains
The subunits themselves are modular [4]and in the major domains are found in layers in the tetrameric structure.
- The 'top' layer is composed of the
.
To help give a better idea of how the glutamate receptor is oriented on the cell surface in the membrane lipid bilayer, as calculated by the Orientations of Proteins in Membranes database(University of Michigan, USA) is shown with the red patch of spheres indicating the boundary of the hydrophobic core closet to the outside of the cell and the dark blue patch of spheres indicating the boundary closest to the inside of the cell.
PDB Entry
3kg2 is a 4 chains structure of sequences from Rattus norvegicus. Full crystallographic information is available from OCA. Although it is billed as the first structure of a full-length glutamate receptor, the carboxy-terminal domain is not present in the structure.
Reference for the structure
- Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009 Dec 10;462(7274):745-56. Epub . PMID:19946266 doi:10.1038/nature08624
Related Structures
- 3kgc GluA2 ligand-binding core complex bound with glutamate
- 2a5t GluN1-GluN2A ligand-binding domain heterodimer
- 2a5s GluN2A ligand-binding domain bound with glutamate
See Also
References
- ↑ Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009 Dec 10;462(7274):745-56. Epub . PMID:19946266 doi:10.1038/nature08624
- ↑ Wollmuth LP, Traynelis SF. Neuroscience: Excitatory view of a receptor. Nature. 2009 Dec 10;462(7274):729-31. PMID:20010675 doi:10.1038/462729a
- ↑ Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009 Dec 10;462(7274):745-56. Epub . PMID:19946266 doi:10.1038/nature08624
- ↑ Wo ZG, Oswald RE. Unraveling the modular design of glutamate-gated ion channels. Trends Neurosci. 1995 Apr;18(4):161-8. PMID:7539962
Page started with original page seeded by OCA on Wed Dec 16 11:24:54 2009 for 3kg2.