T-box proteins
From Proteopedia
(→References) |
(Restoring references - don't know how to do them properly) |
||
| Line 128: | Line 128: | ||
=References= | =References= | ||
| - | + | * Current Opinion in Genetics & Development 1997, 7:474-480; | |
| + | * Gene 258 (2000) 15–29; | ||
| + | * American Journal of Medical Genetics Part A 140A:1407–1413 (2006); | ||
| + | * Genome Biology 2002, 3(6):reviews3008.1–3008.7; | ||
Revision as of 08:28, 7 July 2010
Contents |
Introduction
The family of T-box proteins are transcription factors with a transcriptional activator or repressor domain and a DNA-binding T-box domain. The first two proteins discovered were Brachyury and Omb, which were found to have a new distinct domain unlike any other, designated the T-box domain. Other homologous proteins have been discovered in both invertebrate and vertebrate organisms but not in some organisms such as Aribidopsis thaliana. This family has ~20 members in Homo sapiens.
Gene loci and structure
The loci of these genes are dispersed throughout the genomes of relevant oragnisms, though a few select cases of clustering exist. Tight linkages include Tbx8 and Tbx9 in C. elegans, and in mice: Tbx2 and Tbx4 on chromosome 11; and Tbx3 and Tbx5 on chromosome 5. Similar linkages are present in humans. These genes include multiple exons, typically 8 (as in Tbx5). There are a few cases of alternative splicing, such as VegT in Xenopus and TBX1 in humans.
Protein structure
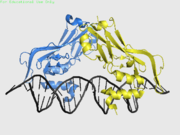
A T-box protein is typically 50 - 78 kDa, within which the T-box domain is typically 17 - 26 kDa. T-box proteins have folds characteristic of the immunoglobin fold. The T-box domains of most family members bind to the consensus sequence TCACACCT. Some residues are conserved through particular subfamilies, such as Lys149 of the Xbra family, and others throughout the entire T-box family. However, differences lie in the binding topology of multiple recognised sequence motifs. For example, Xbra binds two head-to-head sequence motifs, and cannot bind them tail-to-tail; VegT can bind tail-to-tail and not head-to-head. It has been suggested that other functions may lie within the T-box such as mediating protein-protein interactions. The transcription-mediating domain can either be activating or repressing and bears less to no sequence homology.
Function and implications in development
T-box proteins are expressed in specific organs or in only particular cell types, and are involved in decisions about early cell fate. Mutant alleles typically show haploinsufficiency (T-box proteins tend to be dosage-specific). When mutations occur, developmental disorders arise such as the ulnar-mammary syndrome caused by mutations in TBX3, and some aspects of DiGeorge syndrome implicated by mutations in TBX1.
Genes involved in limb bud formation and development are a subset of T-box proteins: T, TBX2, TBX3, TBX5, TBX15 and TBX18.
T-box family members in Homo sapiens
| Members | Notes | Linked diseases |
|---|---|---|
| T | Crystal structure solved | |
| TBX19 |
| Members | Notes | Linked diseases |
|---|---|---|
| TBX1 | Crystal structure solved | DiGeorge syndrome |
| TBX10 | ||
| TBX15 | Cousin syndrome | |
| TBX18 | ||
| TBX20 | ||
| TBX22 | Truncated T-box domain predicted not to bind DNA | Cleft palate with ankyloglossia |
| Members | Notes | Linked diseases |
|---|---|---|
| TBX2 | Mutation of Arg122 destroys DNA binding. Acts as a repressor. TRP-1 promoter is a possible binding site. | Amplified in certain types of breast cancer. |
| TBX3 | Crystal structure solved | Ulnar-mammary syndrome |
| TBX4 | Small patella syndrome | |
| TBX5 | Holt-Oram syndrome |
| Members | Notes | Linked diseases |
|---|---|---|
| TBX6 | ||
| MGA | Also contains leucine zipper domain |
| Members | Notes | Linked diseases |
|---|---|---|
| TBR1 | ||
| EOMES | ||
| TBX21 |
References
- Current Opinion in Genetics & Development 1997, 7:474-480;
- Gene 258 (2000) 15–29;
- American Journal of Medical Genetics Part A 140A:1407–1413 (2006);
- Genome Biology 2002, 3(6):reviews3008.1–3008.7;
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Helen Ginn, Alexander Berchansky, David Canner, Joel L. Sussman