This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Sandbox 39
From Proteopedia
| Line 45: | Line 45: | ||
= Ligands = | = Ligands = | ||
| - | + | A <scene name='Sandbox_39/Ligands_1/1'>ligand</scene> is a protein that is able to bind to the active site of an enzyme to form a biologically relevant complex. Lysozyme reaction is hydrolysis of the beta (1-4) glycosidic bond between N-acetylglucosamine sugar (NAG) and N-acetylmuramic acid sugar (NAM). Lysozyme very specific active site, which can bind only six sugar rings from a polysaccharide chain and hydrolyze them, so these six sugar rings represent the ligand of lysozyme. | |
| - | + | ||
| - | + | ||
Revision as of 22:56, 26 October 2010
| Please do NOT make changes to this Sandbox. Sandboxes 30-60 are reserved for use by Biochemistry 410 & 412 at Messiah College taught by Dr. Hannah Tims during Fall 2012 and Spring 2013. |
Contents |
LYSOZYME
Lysozyme is a powerful enzyme of biological significance found in abundance in tears, saliva, and human milk. It is also known as muramidase, or glycocide hydrolase. It is known for damaging bacterial cell walls by catalyzing the hydrolysis of 1,4-beta-linkages between N-acetylmuramic acid and N-acetyl-D-glucosamine residues in a peptidoglycan and between N-acetyl-D-glucosamine residues in chitodextrins. In this way, lysozyme is efficient in lysing the cell walls of both bacteria and fungi.
A simple cartoon structure of lysozyme can be seen below. The secondary structures can be seen in blue, and the salt bridges are highlighted in yellow. The following sections will highlight different subsections of the lysozyme protein using colors and labels through the program Jmol.
|
Secondary Structure
The structure of lysozyme with its highlighted in yellow and pink can be seen to your left. The structures highlight the alpha helicies, and the yellow lines highlight the beta-pleated sheets. Lysozyme contains five alpha helicies and five beta-pleated sheets. Three of the beta-pleated sheets are antiparallel to one another, and the other two are separate from each other.
Hydrophobicity
Lysozyme contain both hydrophobic, or water-hating, regions and hydrophillic, or water-loving, regions, referred to overall as . These regions can be displayed with the hydrophobic regions in gray and the polar, hydrophillic regions in purple. This coloration highlights the location of these regions, showing the majority of the hydrophobic regions are inside of the protein and the majority of the hydrophillic regions are on the outside of the molecule.
|
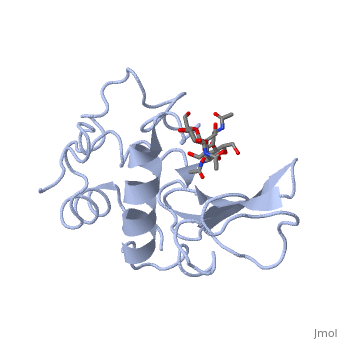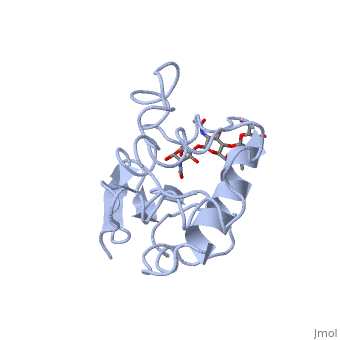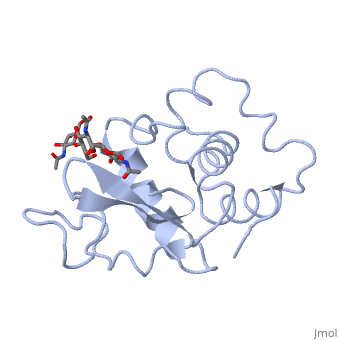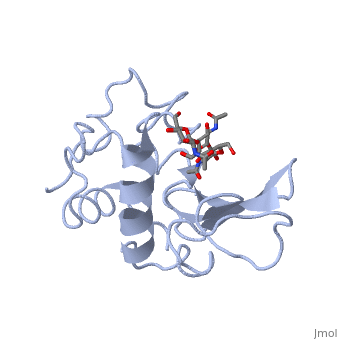
Ligands
A is a protein that is able to bind to the active site of an enzyme to form a biologically relevant complex. Lysozyme reaction is hydrolysis of the beta (1-4) glycosidic bond between N-acetylglucosamine sugar (NAG) and N-acetylmuramic acid sugar (NAM). Lysozyme very specific active site, which can bind only six sugar rings from a polysaccharide chain and hydrolyze them, so these six sugar rings represent the ligand of lysozyme.
Charged Residues
The charged residues, or the most polar portions of the molecule, are seen highlighted to the right in the space-filling model. The blue represent cations, and the red charges represent anions. The can also be seen, with the charged residues the same as above and the polar residues in purple. It is important to note that these residues are found almost exclusively on the outside of the protein to increase its interaction of water.