User:Eran Hodis/Sandbox8
From Proteopedia
Eran Hodis (Talk | contribs)
(New page: ===General Structure=== There are two distinct classes of HMGRs, class I, which is only found in eukaryotes and are membrane bound and class II, which is found in prokaryotes and are solub...)
Next diff →
Revision as of 14:40, 29 October 2010
General Structure
There are two distinct classes of HMGRs, class I, which is only found in eukaryotes and are membrane bound and class II, which is found in prokaryotes and are soluble. [1] HMGR contains 8 transmembrane domains, which have yet to be successfully crystallized, that anchor the protein to the membrane of the endoplasmic reticulum. [2] The catalytic portion of human HMGR forms a tetramer, with the individual monomers winding around each other. [2] Within the tetramer, the monomers are arranged into , each of which contains which are formed by residues form both monomers. Each monomer contains , the , the , and the . The L-domain is unique to HMGRs while the S-domain, which forms the binding site for NADP, resembles that of ferredoxin. The S and L domains are connected by a which is essential for the HMG-binding site. [2] Salt bridges between residues R641 and E782 as well as between E700 and E700 on neighboring monomers compliment the largely hydrophobic dimer-dimer interface. [2]
Substrate Binding & Catalytic Mechanism
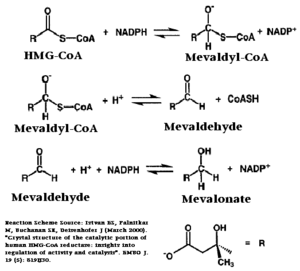
The HMG-CoA and NADPH molecules make numerous contacts with the L and S domains in forming the four active sites. The CoA is located in a , with the pantothenic acid moiety extending into the interior of the protein. over the CoA adenine base, shielding the extended binding pocket from solution. The NADPH binding site is formed primarily by the S-domain with playing a critical role in binding. [2]
The HMG binding pocket is the site of catalysis in HMGR. is a critical structural element of this binding site. Residues and are positioned in the active site as is . It is this K691 that presumably stabilizes the negatively charged oxygen on the first mevaldyl-CoA intermediate. [2] The mevaldyl CoA intermediate is subsequently converted to Mavaldehyde with added stabilization from . It is then believed that the close proximity of increases the pKA of E559, allowing it to be a proton donor for the reduction of mevaldehyde into mevalonate. [2]