We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Eran Hodis/Sandbox3
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| - | ==Your Heading Here (maybe something like 'Structure')== | ||
| - | <StructureSection load='1dq8' size='600' side='right' caption='Structure of HMG-CoA reductase (PDB entry [[1dq8]])'>test</StructureSection> | ||
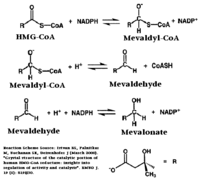
==Structural Features== | ==Structural Features== | ||
Revision as of 12:10, 1 November 2010
Structural Features
| |||||||||||
|
References
- ↑ Istvan ES, Deisenhofer J. Structural mechanism for statin inhibition of HMG-CoA reductase. Science. 2001 May 11;292(5519):1160-4. PMID:11349148 doi:10.1126/science.1059344
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 Roitelman J, Olender EH, Bar-Nun S, Dunn WA Jr, Simoni RD. Immunological evidence for eight spans in the membrane domain of 3-hydroxy-3-methylglutaryl coenzyme A reductase: implications for enzyme degradation in the endoplasmic reticulum. J Cell Biol. 1992 Jun;117(5):959-73. PMID:1374417
- ↑ 3.0 3.1 Meigs TE, Roseman DS, Simoni RD. Regulation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase degradation by the nonsterol mevalonate metabolite farnesol in vivo. J Biol Chem. 1996 Apr 5;271(14):7916-22. PMID:8626470
- ↑ Song BL, Sever N, DeBose-Boyd RA. Gp78, a membrane-anchored ubiquitin ligase, associates with Insig-1 and couples sterol-regulated ubiquitination to degradation of HMG CoA reductase. Mol Cell. 2005 Sep 16;19(6):829-40. PMID:16168377 doi:10.1016/j.molcel.2005.08.009
- ↑ Goldstein JL, Brown MS. Regulation of the mevalonate pathway. Nature. 1990 Feb 1;343(6257):425-30. PMID:1967820 doi:http://dx.doi.org/10.1038/343425a0