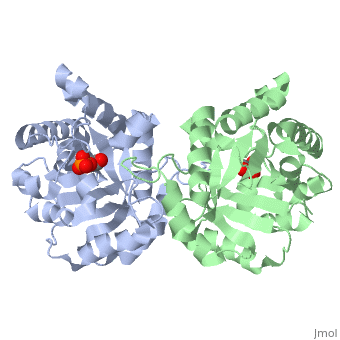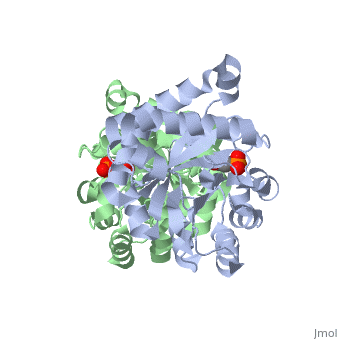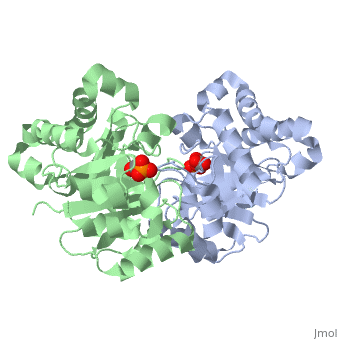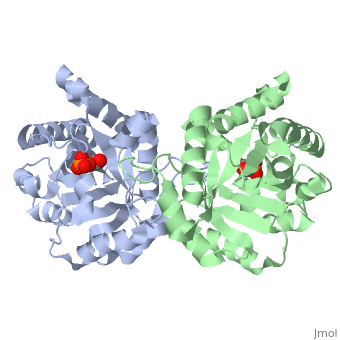From Proteopedia
(Difference between revisions)
proteopedia linkproteopedia link
|
|
| Line 42: |
Line 42: |
| | | | |
| | == Structure & Function == | | == Structure & Function == |
| - | <Structure load='2ypi' size='300' frame='true' align='right'/> | + | <Structure load='2ypi' size='300' frame='true' align='right' scene='Triose_Phosphate_Isomerase/Helix_shaded_sheet_3/1'>alpha helices'/> |
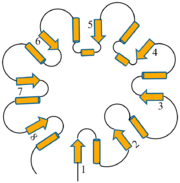
| | Triose Phosphate Isomerase is a member of the all alpha and beta (α/β) class of proteins and it is a homodimer consisting of two nearly identical subunits each consisting of 247 amino acids and differing only at their N-terminal ends. Each TPI monomer contains the full set of catalytic residues; however, the enzyme is only active in the oligomeric form. <ref>PMID:18562316</ref> Therefore, dimerization is essential for full function of the enzyme even though it is not believed that any cooperativity exists between the two active sites.<ref>PMID: 2065677</ref> Each subunit contains 8 exterior <scene name='Triose_Phosphate_Isomerase/Helix_shaded_sheet_3/1'>alpha helices</scene> surrounding 8 interior <scene name='Triose_Phosphate_Isomerase/Beta_sheet_labelled/1'>beta sheets</scene>, which form a conserved structural domain called a closed alpha/beta barrel (αβ) or more specifically a <scene name='Triose_Phosphate_Isomerase/Tim_barrel_2/1'>TIM Barrel</scene>. The TIM barrel was originally named after TPI and is estimated to be present in 10% of all enzymes. Characteristic of most all TIM barrel domains is the presence of the enzyme's active site in the lower loop regions created by the eight loops that connect the C-terminus of the beta strands with the N-terminus of the alpha helices. TIM barrel proteins also share a structurally conserved phosphate binding motif, with the phosphate either coming from the substrate or from cofactors. <ref> http://www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv</ref>. | | Triose Phosphate Isomerase is a member of the all alpha and beta (α/β) class of proteins and it is a homodimer consisting of two nearly identical subunits each consisting of 247 amino acids and differing only at their N-terminal ends. Each TPI monomer contains the full set of catalytic residues; however, the enzyme is only active in the oligomeric form. <ref>PMID:18562316</ref> Therefore, dimerization is essential for full function of the enzyme even though it is not believed that any cooperativity exists between the two active sites.<ref>PMID: 2065677</ref> Each subunit contains 8 exterior <scene name='Triose_Phosphate_Isomerase/Helix_shaded_sheet_3/1'>alpha helices</scene> surrounding 8 interior <scene name='Triose_Phosphate_Isomerase/Beta_sheet_labelled/1'>beta sheets</scene>, which form a conserved structural domain called a closed alpha/beta barrel (αβ) or more specifically a <scene name='Triose_Phosphate_Isomerase/Tim_barrel_2/1'>TIM Barrel</scene>. The TIM barrel was originally named after TPI and is estimated to be present in 10% of all enzymes. Characteristic of most all TIM barrel domains is the presence of the enzyme's active site in the lower loop regions created by the eight loops that connect the C-terminus of the beta strands with the N-terminus of the alpha helices. TIM barrel proteins also share a structurally conserved phosphate binding motif, with the phosphate either coming from the substrate or from cofactors. <ref> http://www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv</ref>. |
| | [[Image:beta barrel.png|left|thumb|'''TIM Barrel''']] | | [[Image:beta barrel.png|left|thumb|'''TIM Barrel''']] |
Revision as of 14:03, 6 November 2010
Template:STRUCTURE 2ypi
Triose Phosphate Isomerase (TPI or TIM) [5.3.1.1] is a ubiquitous dimeric enzyme with a molecular weight of roughly 54 kD (27 kD per subunit) which catalyzes the reversible interconversion of the triose phosphate isomers dihydroxyacetone phosphate (DHAP) and D-glyceraldehyde-3-phosphate (GAP), an essential process in the glycolytic pathway. More simply, the enzyme catalyzes the isomerization of a ketose (DHAP) to an aldose GAP also referred to as PGAL. In regards to the two isomers, at equilibrium, roughly 96% of the triose phosphate is in the DHAP isomer form; however, the isomerization reaction proceeds due to the rapid removal of GAP from the subsequent reactions of glycolysis. The TPI structure is shown on the right (PDB entry 2ypi) in complex with GAP, which is bound to each of its two active sites. TPI is an example of a catalytically perfect enzyme, indicating that for almost every enzyme-substrate encounter, a product is formed and that this interaction is limited only by the substrate diffusion rate. In addition to its role in glycolysis, TPI is also involved in several additional metabolic biological processes including gluconeogenesis, the pentose phosphate shunt, and fatty acid biosynthesis. A point mutation to a glutamate residue (Glu104) of TPI results in triose phosphate isomerase deficiency, an autosomal recessive inherited disorder characterized by an increased accumulation of DHAP in erythrocytes. Structurally, this point mutation abolishes TPI’s ability to dimerize, subsequently inhibiting its catalytic activity.
Mechanism
TPI catalyzes the transfer of a hydrogen atom from carbon 1 to carbon 2, an intramolecular oxidation-reduction reaction

Isomerization reaction catalyzed by TPI . This isomerization of a ketose to an aldose proceeds through an
cis-enediol(ate) intermediate. This isomerization proceeds without any cofactors and the enzyme confers a 10
9 rate enhancement relative to the nonenzymatic reaction involving a chemical base (acetate ion).
[1].

Kinetic constants of Triose Phosphate Isomerase Acid-Base Catalysis
The mechanism of TPI has been extensively studied by prominent enzymologists for several decades leading to several different proposed mechanisms of catalysis. The original "Classic" mechanism put forth by Knowles and co-workers is outlined in the mechanism provided below.[2]
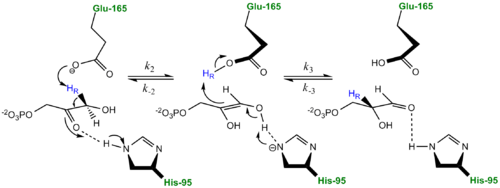
Classic Mechanism proposed by Knowles and co-workers TPI carries out the isomerization reaction through an acid-base-mediated mechanism involving . First the DHAP or GAP subtrate is initially attracted to the enzyme active site through electrostatic interactions between the negatively charged substrate phosphate group and the positively charged , with the resulting interaction stabilizing the substrate. According to the "classic" mechanism, plays the role of the general base catalyst by abstracting a proton from the pro(R) position of carbon 1 of DHAP or the C-2 proton of GAP. However, the carboxylate group of Glutamate 165 alone does not possess the basicity to abstract a proton and requires , the general acid, to donate a proton to stabilize the negative charge building up on C-2 carbonyl oxygen, effectively stabilizing the planar endediol(ate) intermediate,. Lys12 and Asn11 also function to stabilize the negative charge which builds up on this intermediate. At this point in the mechanism, Glutamate 165 acts as a general acid by donating its proton to the neighboring C-2, while Histidine 95 now acts as a general base by abstracting a proton from the hydroxyl group of C-1. The final step in the reaction is the formation of the GAP isomer product while glutamate and histidine are returned to their original forms, regenerating the enzyme. In studies using tritium labeled DHAP, Knowles observed only ~ 6% intramolecular transfer of the 3H label to the GAP product. In explaining this result, Knowles argued that the hydrogen bound to the Glu165 was in equilibrium with those in bulk solvent. Additionally, the reaction mechanism of the methylglyoxal forming enzyme methylglyoxal synthase (MGS) is believed to be similar to that of triosephosphate isomerase. Both enzymes utilize DHAP to form an enediol(ate) phosphate intermediate as the first step of their reaction pathways; however, the second catalytic step in the MGS reaction pathway features the elimination of phosphate and collapse of the enediol(ate) to form methylglyoxal rather then reprotonation to form the isomer glyceraldehyde 3-phosphate as seen in TPI.[3]
The Enediol(ate) Intermediate as a Kinetic Barrier
The formation and stabilization of the enediol(ate) intermediate has been a source of great discussion amongst scientists in the field. This is due to the fact that the formation of the enediol(ate) intermediate presents a large thermodynamic barrier involving the abstraction of the α-proton from the carbon acid substrate, an unfavorable process due to the lack of acidity and high pKa of the C1 proton of the substrate. The "classic" mechanism (shown above) put forth by Knowles and co-workers stated that the kinetic barrier is overcome by the concerted deprotonation of the C1 and the protonation of the C2 carbonyl oxygen which would effectively allow for equivalent free energies for each species shown shown, thus promoting an equilibrium constant near unity.[4] This concept of preferential binding arising from matching of the reaction energy barriers is a common theme of enzyme catalysis [5].
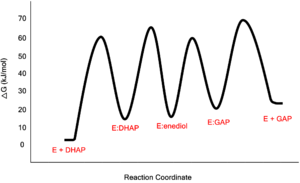
Free-Energy Profile for the Reaction Catalyzed by Triosephosphate Isomerase Low-Barrier Hydrogen Bond in the TPI Mechanism
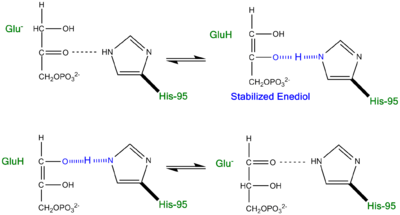
Proposed LBHB between Histidine and Enediol Intermediate.
An additional explanation of the TPI mechanism proposed by Cleeland and Kreevoy stipulates the formation of a Low-barrier hydrogen bond[6]. Support for this LBHB arose from the rare observation of a hydrogen bond between the carbonyl oxygen of the substrate and a neutral histidine. It was reasoned that a neutral histidine was required to match the pKa of the enediol, a requirement for the formation of a shorter and stronger LBHB (pKa's ~ 14). It was rationalized that this strengthened hydrogen bond and ideal geometry would effectively speed up the enolization reaction. Structural evidence for this LBHB was found in a 1.2 Å crystal structure of TIM complexed with DHAP demonstrating an extremely short hydrogen bond (2.6 Å) between His95 and O2 of DHAP [7]. Under the mechanism stipulating a LBHB between His95 and O2 of DHAP, Glu165 would catalyze all proton transfers between C1 and C2, while His95 would act as an electrophilic catalyst by forming a close, stabilizing LBHB with the cis-enediolate intermediate.
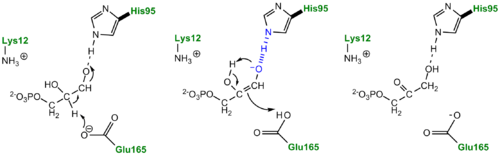
TPI Mechanism with LBHB between His95 and O2 of substrate More recently a series of NMR experiments carried out by Mildvan and co-workers have shed light onto an alternative "Criss-cross" mechanism involving a LBHB between the catalytic Glu165 and the O1 oxygen of the substrate. This mechanism stipulates the His95 side chain does not directly transfer protons, this rather being accomplished entirely by Glu165. Support for this mechanism was provided by Richard and coworkers who carried tritium labeling experiments demonstrating a significant amount of intramolecular transfer (49%) of the 1H label from substrate (DHAP) to product (GAP)[8]. Using phosphoglycolohydroxamate (PGH), a mimic of the enediol(ate) intermediate, a 14.9 ppm chemical shift (6 ppm downfield) as well as a deuterium fractionation factor of 0.38 was observed with the TIM-PGH complex, corresponding to a highly deshielded proton involved in a LBHB between Glu165 and the hydroxamate oxygen of PGH. Conversely, the same NMR study found an additional hydrogen bond between the N-ε proton of His95 and the carbonyl oxygen of PGH; however, its chemical shift of 13.5 (0.4 ppm downfield from free enzyme) and fractionation factor of 0.71 indicated this was a strong H-bond, but not a LBHB.[9].
The formation of the LBHB between Glu165 and O1 of the inhibitor PGH is due to the matching of p
Kas and the alternative mechanism suggests that Glu-165, in addition to its role in initially abstracting the proton from the substrate, may also shuttle protons to and from the oxygens in the intermediate. Also, the "criss-cross" mechanism implies that the by donating a normal hydrogen bond the role of His95 is to polarize the carbonyl oxygen and lower the p
Ka of PGH in order to facilitate subsequent proton abstraction
[10]. It has been argued that that the LBHB formed between Glu165 and PGH is a consequence of using the inhibitor PGH, whose hydroxamate p
Ka of 9 better matches Glu165 then His95, and that the biological reaction would instead see the enediol forming a LHBH with His95, as mentioned above. Overall, the mechanism employed by TPI has yet to be completely solved and recent NMR studies involving both WT and mutant TPI enzymes have revealed contributions from both the "classic" and "criss-cross" mechanisms.
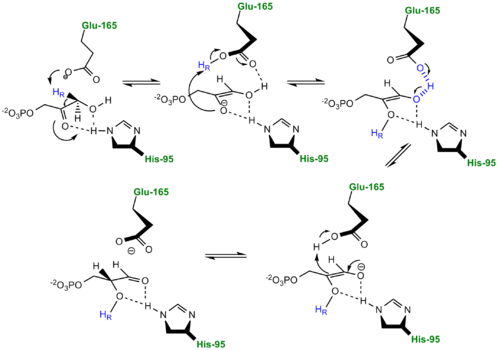
Alternative "Criss-Cross" TPI Mechanism Involving LBHB Between Glu165 and O1 of the Intermediate Inhibitors of Triose Phosphate Isomerase
Although a highly studied enzyme, there are relatively few effective inhibitors of TPI. From a pharmaceutical perspective, if TPI structures differ greatly between humans and microorganisms such as Plasmodium or Trypanosoma, whose growth rely heavily or entirely on glycolysis, inhibition may be a strong therapeutic target.[11] Two irreversible inhibitors, halo-acetone phosphate and glycidol phosphate (1,2-epoxypropanol-3-P), act by labeling active site residues. Early biochemical studies involving glycidol phosphate have revealed the labeled residue to be the active site glutamate. There are several weak reversible inhibitors of TPI including 3-Phosphoglycerate, glycerol phosphate and phosphoenol pyruvate, with Ki values ranging from 0.2-1.3 mM.[12] Additionally, several transition state analogues have been used to study the mechanism of TPI, including phosphoglycolohydroxamate (Ki = 6-14 μM) and the phosphoglycolic acid
(Ki = 3 μM) and 2(N-formyl-N-hydroxy)aminoethyl phosphonate (IPP) [13]. PGA (also called 2PG) believed to bind TPI as a trianion, undergoes tight active site binding through electrostatic interactions with both the neutral His95 and protonated Glu165 side chains. PGH (binding in the cis conformation) and IPP function by mimicking structural features of the cognate DHAP and GAP substrates, respectively[14]. Specifically, PGH effectively mimics the planar enediol(ate)intermediate.
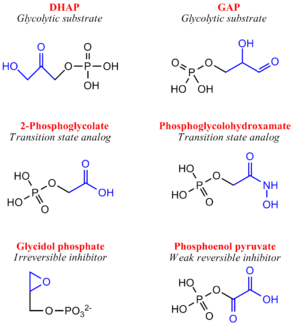
Inhibitors of Triose Phosphate Isomerase Structure & Function
| alpha helices'/>
Triose Phosphate Isomerase is a member of the all alpha and beta (α/β) class of proteins and it is a homodimer consisting of two nearly identical subunits each consisting of 247 amino acids and differing only at their N-terminal ends. Each TPI monomer contains the full set of catalytic residues; however, the enzyme is only active in the oligomeric form. [15] Therefore, dimerization is essential for full function of the enzyme even though it is not believed that any cooperativity exists between the two active sites.[16] Each subunit contains 8 exterior surrounding 8 interior , which form a conserved structural domain called a closed alpha/beta barrel (αβ) or more specifically a . The TIM barrel was originally named after TPI and is estimated to be present in 10% of all enzymes. Characteristic of most all TIM barrel domains is the presence of the enzyme's active site in the lower loop regions created by the eight loops that connect the C-terminus of the beta strands with the N-terminus of the alpha helices. TIM barrel proteins also share a structurally conserved phosphate binding motif, with the phosphate either coming from the substrate or from cofactors. [17].
Ω Loop 6
As mentioned earlier, TPI is considered a catalytically perfect enzyme and accomplishes this largely due to its ability to suppress or prevent undesired side reactions such as the decomposition of the enediol intermediate into methyl glyoxal and orthophosphate, a process which is 100 fold faster in solution than the desired isomerization. TPI is able to prevent this undesired reaction by trapping and stabilizing the charged endiol(ate) intermediate in the active site through the use of a flexible 11 residue Ω loop referred to as containing residues 168-178[18], residue numbers variable with regards to species. Loop 6 can be further divided into a 3-residue N-terminal hinge, a rigid hydrophobic lid spanning 5-residues and a 3-residue C-terminal hinge . The complete closure of this loop, a movement of roughly 7 Å for the tip of the loop (Cα of Thr172) and occurring on a microsecond timescale, is facilitated by several hydrogen bonding interactions between loop 6 and loop 7 including H-bonds between the hydroxyl group of Tyrosine 208 (loop 7) and the amine nitrogen of Alanine 176 as well as between Serine 211 (loop 7) and Glycine 173. As mentioned above, the loop shuts when the enediol is present, effectively shielding both ligand and catalytic residues from solvent exposure, and reopens when the isomerization is complete. Site-directed mutagenesis experiments substituting a Phenylalanine for the Tyrosine resulted in a 2400-fold decrease in catalytic activity. [19] and it is beleived the opening/closing of loop 6 and loop 7 is partially rate-limiting. Additionally, extensive mechanistic and kinetic experiments involving Trypanosoma brucei, a parasitic protist causing sleeping sickness in humans, has revealed the structural and functional importance of a proline residue at position 168 in conjunction with transmitting the signal of ligand binding to the conformational change of the catalytic glutamate residue (Glu167 in T.brucei) and the subsequent proper loop 6 closure.[20] Specifically, the proline residue is positioned at the beginning of loop 6 as to aid in the catalytic glutamate side chain flipping from the inactive swung-out to the active swung-in conformation, facilitating the closure of the loop. Structurally, in the unliganded (open) conformation, the Glu-Pro peptide bond is in the energetically favored trans conformation; however, in the liganded (closed) conformation, the pyrrolidine ring of proline adopts a rare strained planar conformation (9 kJ/mol in vacuo), suggesting that the strain could be important for loop opening and product release, upon completion of the reaction cycle.[21]
Entropic Effects of Ω Loop 6 Hinges
Similar to the loop spanning residues, the Ω loop 6 hinge residues share high sequence homology amongst species. The role of both the N- and C-terminal hinge regions of Ω loop 6 have been extensively studied including the replacement of conserved hinge residues with glycine, which resulted in a 2500-fold drop in kcat. The insertion of glycine into the hinge region significantly increases the flexibility of the loop due to glycine's conformational freedom, which in turn allows the loop to sample many more conformations. This has thermodynamic ramifications as these glycine-rich hinge mutants prompted a large entropy gain (+ΔS) compared to WT, effectively altering the entropic activation energy. Specifically, WT TPI is able to overcome the initial entropic gain (order to disorder), caused by dispelling water molecules from the active site, by forming a more ordered enzyme-substrate complex. Conversely, the glycine-rich hinge mutants again promote an initial entropy gain due to water loss but are unable to pay the entropic penalty due to their inability to bind substrate tightly. Since catalysis will only occur when the closed conformation has been sampled it reasons that the likelihood of sampling this conformation is greatly reduced with the glycine-rich hinge mutants, as evidence by a significant drop in kcat. As their overall biological role in the enzyme, the loop hinges act to limit the motion of the loop which effectively restricts the number of conformations accessible to the enzyme. In this manner, TPI acts like an entropy trap.
Disease
Triose Phosphate Isomerase Deficiency: Initially described in 1965, TPI deficiency is an autosomal recessive inherited disorder with characteristics ranging from chronic haemolytic anaemia, increased susceptibility to infections, severe neurological dysfunction, and often times death in early childhood.[22] TPI has been most closely linked to a point mutation at the residue which results in the mutation. A common marker for TPI deficiency is the increased accumulation of dihydroxyacetone phosphate in erythrocyte extracts as a result in the inability of the mutant enzyme to catalyze the isomerization to D-glyceraldehyde-3-phosphate. Recent evidence has indicated that the point mutation does not prove detrimental to the rate of catalysis of the enzyme, but rather effects the ability of the enzyme to dimerize.[23]
Role in Alzheimer's Disease: Recent discoveries in Alzheimer Disease research has indicated that amyloid beta-peptide induced nitro-oxidative damage promotes the nitrotyrosination of the glycolytic enzyme triosephosphate isomerase in human neuroblastoma cells.[24] nitro-triosephosphate isomerase was found to be present in brain slides from double transgenic mice overexpressing human amyloid precursor protein as well as in Alzheimer's disease patients. Specifically, the nitrotyrosination occurs on , which are located in close proximity to the catalytic center, and this modification correlates with a reduced isomerase activity. Additionally, according to work done by Francesc Guix and colleagues, nitro-triosphosphate isomerase contributed to the formation of large beta-sheet aggregates in vitro and in vivo.
Evolutionary Conservation
Due to its role in the glycolysis, an essential process to many organisms, TPI has been isolated and crystallized from several species giving rise to extensive multiple alignment in silico experiments which subsequently provided of TPI. [25] Collectively, these tools have determined that TPI has a roughly 50% sequence conservation from bacteria to humans.[26] One specific example of sequence homology is that of loop 6 and loop 7 residues, whose structural contributions are discussed above. In a sequence alignment of 133 TIM sequences, two highly conserved motifs are noticed. First, 114 sequences in loop 6 contain the PXW sequence family (where X is I,L or V in 112 sequences or otherwise a T or K). Secondly, loop 7 contains a highly conserved YGGS motif; however, this motif is only found when the N-terminal hinge contains tryptophan.
Solved TPI Structures
- 1tim - TPI - Gallus gallus
- 2ypi - TPI + PGA - Saccharomyces cerevisiae
- 2vom - TPI Glu104Asp mutant - Homo sapiens
- 1wyi - TPI - Homo sapiens
- 2j27 - TPI Pro168Ala mutant -Trypanosoma brucei
Additional Resources
For additional information, see: Carbohydrate Metabolism
References
- ↑ Davenport RC, Bash PA, Seaton BA, Karplus M, Petsko GA, Ringe D. Structure of the triosephosphate isomerase-phosphoglycolohydroxamate complex: an analogue of the intermediate on the reaction pathway. Biochemistry. 1991 Jun 18;30(24):5821-6. PMID:2043623
- ↑ Harris TK, Abeygunawardana C, Mildvan AS. NMR studies of the role of hydrogen bonding in the mechanism of triosephosphate isomerase. Biochemistry. 1997 Dec 2;36(48):14661-75. PMID:9398185 doi:10.1021/bi972039v
- ↑ Saadat D, Harrison DH. The crystal structure of methylglyoxal synthase from Escherichia coli. Structure. 1999 Mar 15;7(3):309-17. PMID:10368300
- ↑ Harris TK, Abeygunawardana C, Mildvan AS. NMR studies of the role of hydrogen bonding in the mechanism of triosephosphate isomerase. Biochemistry. 1997 Dec 2;36(48):14661-75. PMID:9398185 doi:10.1021/bi972039v
- ↑ Rozovsky S, McDermott AE. Substrate product equilibrium on a reversible enzyme, triosephosphate isomerase. Proc Natl Acad Sci U S A. 2007 Feb 13;104(7):2080-5. Epub 2007 Feb 7. PMID:17287353 doi:http://dx.doi.org/0608876104
- ↑ Cleland WW, Kreevoy MM. Low-barrier hydrogen bonds and enzymic catalysis. Science. 1994 Jun 24;264(5167):1887-90. PMID:8009219
- ↑ Jogl G, Rozovsky S, McDermott AE, Tong L. Optimal alignment for enzymatic proton transfer: structure of the Michaelis complex of triosephosphate isomerase at 1.2-A resolution. Proc Natl Acad Sci U S A. 2003 Jan 7;100(1):50-5. Epub 2002 Dec 30. PMID:12509510 doi:10.1073/pnas.0233793100
- ↑ O'Donoghue AC, Amyes TL, Richard JP. Hydron transfer catalyzed by triosephosphate isomerase. Products of isomerization of (R)-glyceraldehyde 3-phosphate in D2O. Biochemistry. 2005 Feb 22;44(7):2610-21. PMID:15709774 doi:10.1021/bi047954c
- ↑ Cleland WW, Frey PA, Gerlt JA. The low barrier hydrogen bond in enzymatic catalysis. J Biol Chem. 1998 Oct 2;273(40):25529-32. PMID:9748211
- ↑ Cleland WW, Frey PA, Gerlt JA. The low barrier hydrogen bond in enzymatic catalysis. J Biol Chem. 1998 Oct 2;273(40):25529-32. PMID:9748211
- ↑ Fonvielle M, Mariano S, Therisod M. New inhibitors of rabbit muscle triose-phosphate isomerase. Bioorg Med Chem Lett. 2005 Jun 2;15(11):2906-9. PMID:15911278 doi:10.1016/j.bmcl.2005.03.061
- ↑ Fonvielle M, Mariano S, Therisod M. New inhibitors of rabbit muscle triose-phosphate isomerase. Bioorg Med Chem Lett. 2005 Jun 2;15(11):2906-9. PMID:15911278 doi:10.1016/j.bmcl.2005.03.061
- ↑ Fonvielle M, Mariano S, Therisod M. New inhibitors of rabbit muscle triose-phosphate isomerase. Bioorg Med Chem Lett. 2005 Jun 2;15(11):2906-9. PMID:15911278 doi:10.1016/j.bmcl.2005.03.061
- ↑ Kursula I, Wierenga RK. Crystal structure of triosephosphate isomerase complexed with 2-phosphoglycolate at 0.83-A resolution. J Biol Chem. 2003 Mar 14;278(11):9544-51. Epub 2003 Jan 9. PMID:12522213 doi:http://dx.doi.org/10.1074/jbc.M211389200
- ↑ Rodriguez-Almazan C, Arreola R, Rodriguez-Larrea D, Aguirre-Lopez B, de Gomez-Puyou MT, Perez-Montfort R, Costas M, Gomez-Puyou A, Torres-Larios A. Structural basis of human triosephosphate isomerase deficiency: mutation E104D is related to alterations of a conserved water network at the dimer interface. J Biol Chem. 2008 Aug 22;283(34):23254-63. Epub 2008 Jun 18. PMID:18562316 doi:10.1074/jbc.M802145200
- ↑ Schnackerz KD, Gracy RW. Probing the catalytic sites of triosephosphate isomerase by 31P-NMR with reversibly and irreversibly binding substrate analogues. Eur J Biochem. 1991 Jul 1;199(1):231-8. PMID:2065677
- ↑ http://www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv
- ↑ Joseph D, Petsko GA, Karplus M. Anatomy of a conformational change: hinged "lid" motion of the triosephosphate isomerase loop. Science. 1990 Sep 21;249(4975):1425-8. PMID:2402636
- ↑ Derreumaux P, Schlick T. The loop opening/closing motion of the enzyme triosephosphate isomerase. Biophys J. 1998 Jan;74(1):72-81. PMID:9449311 doi:10.1016/S0006-3495(98)77768-9
- ↑ Casteleijn MG, Alahuhta M, Groebel K, El-Sayed I, Augustyns K, Lambeir AM, Neubauer P, Wierenga RK. Functional role of the conserved active site proline of triosephosphate isomerase. Biochemistry. 2006 Dec 26;45(51):15483-94. Epub 2006 Dec 19. PMID:17176070 doi:10.1021/bi061683j
- ↑ Kursula I, Wierenga RK. Crystal structure of triosephosphate isomerase complexed with 2-phosphoglycolate at 0.83-A resolution. J Biol Chem. 2003 Mar 14;278(11):9544-51. Epub 2003 Jan 9. PMID:12522213 doi:http://dx.doi.org/10.1074/jbc.M211389200
- ↑ Schneider AS. Triosephosphate isomerase deficiency: historical perspectives and molecular aspects. Baillieres Best Pract Res Clin Haematol. 2000 Mar;13(1):119-40. PMID:10916682
- ↑ Ralser M, Heeren G, Breitenbach M, Lehrach H, Krobitsch S. Triose phosphate isomerase deficiency is caused by altered dimerization--not catalytic inactivity--of the mutant enzymes. PLoS ONE. 2006 Dec 20;1:e30. PMID:17183658 doi:10.1371/journal.pone.0000030
- ↑ Guix FX, Ill-Raga G, Bravo R, Nakaya T, de Fabritiis G, Coma M, Miscione GP, Villa-Freixa J, Suzuki T, Fernandez-Busquets X, Valverde MA, de Strooper B, Munoz FJ. Amyloid-dependent triosephosphate isomerase nitrotyrosination induces glycation and tau fibrillation. Brain. 2009 May;132(Pt 5):1335-45. Epub 2009 Feb 27. PMID:19251756 doi:10.1093/brain/awp023
- ↑ Parthasarathy S, Ravindra G, Balaram H, Balaram P, Murthy MR. Structure of the Plasmodium falciparum triosephosphate isomerase-phosphoglycolate complex in two crystal forms: characterization of catalytic loop open and closed conformations in the ligand-bound state. Biochemistry. 2002 Nov 5;41(44):13178-88. PMID:12403619
- ↑ Joseph-McCarthy D, Lolis E, Komives EA, Petsko GA. Crystal structure of the K12M/G15A triosephosphate isomerase double mutant and electrostatic analysis of the active site. Biochemistry. 1994 Mar 15;33(10):2815-23. PMID:8130194
|