Guanine-Binding Riboswitch
From Proteopedia
| Line 21: | Line 21: | ||
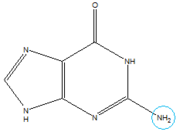
In the experiment described by the abstract above, hypoxanthine binds to the junction via a series of hydrogen bonds with nucleotides <scene name='1u8d/Active_site_hpa/2'>U22, U47, U51 and C74</scene>. The mRNA contacts all of the functional groups in hypoxanthine, which explains its specificity as a ligand. Additionally, the two carbonyl oxygens located at the 2-position of <scene name='1u8d/Guanine_binding_carbonyls/1'>C74 and U51</scene> are able to form hydrogen bonds with guanine's exocyclic amino group [[Image:Guanine_Structure.png|thumb|left|Structure of Guanine with exocyclic amino group circled in blue.]]. This gives the riboswitch a tenfold higher affinity for guanine over hypoxanthine <ref name=bateyetal/>. | In the experiment described by the abstract above, hypoxanthine binds to the junction via a series of hydrogen bonds with nucleotides <scene name='1u8d/Active_site_hpa/2'>U22, U47, U51 and C74</scene>. The mRNA contacts all of the functional groups in hypoxanthine, which explains its specificity as a ligand. Additionally, the two carbonyl oxygens located at the 2-position of <scene name='1u8d/Guanine_binding_carbonyls/1'>C74 and U51</scene> are able to form hydrogen bonds with guanine's exocyclic amino group [[Image:Guanine_Structure.png|thumb|left|Structure of Guanine with exocyclic amino group circled in blue.]]. This gives the riboswitch a tenfold higher affinity for guanine over hypoxanthine <ref name=bateyetal/>. | ||
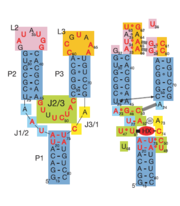
| - | Binding of the ligand causes the RNA to form a three-dimentional fold in which the terminal loops <scene name='Guanine-Binding_Riboswitch/Terminal_loop_l2/ | + | Binding of the ligand causes the RNA to form a three-dimentional fold in which the terminal loops <scene name='Guanine-Binding_Riboswitch/Terminal_loop_l2/4'>L2</scene> and <scene name='Guanine-Binding_Riboswitch/Terminal_loop_l3/3'>L3</scene> become connected through a series of hydrogen bonds (Fig. 1). This causes the <scene name='Guanine-Binding_Riboswitch/Helix_p2/1'>P2</scene> and <scene name='Guanine-Binding_Riboswitch/Helix_p3/1'>P3</scene> helices to become parallel to each other. The unfavorable electrostatic interactions of the parallel ribose-phosphate backbones are neutralized through the binding multiple Co(NH<sub>3</sub>)<sub>6</sub><sup>3+</sup> <scene name='Guanine-Binding_Riboswitch/Cobalt_cations/1'>cations</scene> between the backbones<ref name=bateyetal/>. |
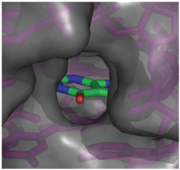
When bound, the ligand is almost completely enveloped by the RNA (Fig. 2). [[Image:Hypoxanthine_enveloped_by_RNA.png|thumb|'''Fig. 2:''' Molecular surface representation of the binding pocket of the guanine riboswitch bound to hypoxanthine.]] This implies that the binding site must undergo significant conformational change upon ligand binding, as a preformed binding site would inhibit the ligand's access to it <ref name=bateyetal/>. | When bound, the ligand is almost completely enveloped by the RNA (Fig. 2). [[Image:Hypoxanthine_enveloped_by_RNA.png|thumb|'''Fig. 2:''' Molecular surface representation of the binding pocket of the guanine riboswitch bound to hypoxanthine.]] This implies that the binding site must undergo significant conformational change upon ligand binding, as a preformed binding site would inhibit the ligand's access to it <ref name=bateyetal/>. | ||
Revision as of 06:49, 28 November 2010
Template:ABSTRACT PUBMED 15549109
Riboswitches
Riboswitches are genetic regulatory elements found in mRNA, where they bind small molecules and directly control gene expression in the absence of regulatory proteins[1]. Most contain a single binding site that recognizes a specific ligand. The ability of a riboswitch to discriminate against molecules that are similar or closely related to its ligand is essential to prevent metabolic misregulation[2].
Evidence that RNA behaves this way comes from studying synthetic binding sites that have been engineered to bind to many ligands, with both high affinity and specificity. While some of these artificial binding sites are used in cells as designer gene control elements, they tend to be smaller, weaker-binding, and less selective than natural binding sites[2].
Atomic-resolution structures of riboswitch binding sites show that they make numerous hydrogen bonds with their ligands, forming contacts that stabilize RNA interactions to further increase affinity. Some binding sites form pockets that entirely engulf the ligand, and in these instances an induced-fit mechanism of binding must occur[2].
Structure
The structure of this riboswitch consists of three helices, and , that surround a .
In the experiment described by the abstract above, hypoxanthine binds to the junction via a series of hydrogen bonds with nucleotides . The mRNA contacts all of the functional groups in hypoxanthine, which explains its specificity as a ligand. Additionally, the two carbonyl oxygens located at the 2-position of are able to form hydrogen bonds with guanine's exocyclic amino group . This gives the riboswitch a tenfold higher affinity for guanine over hypoxanthine [1].Binding of the ligand causes the RNA to form a three-dimentional fold in which the terminal loops and become connected through a series of hydrogen bonds (Fig. 1). This causes the and helices to become parallel to each other. The unfavorable electrostatic interactions of the parallel ribose-phosphate backbones are neutralized through the binding multiple Co(NH3)63+ between the backbones[1].
When bound, the ligand is almost completely enveloped by the RNA (Fig. 2).Reference
- ↑ 1.0 1.1 1.2 1.3 Batey RT, Gilbert SD, Montange RK. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004 Nov 18;432(7015):411-5. PMID:15549109 doi:10.1038/nature03037
- ↑ 2.0 2.1 2.2 Breaker, Ronald R. (28 March, 2008). Complex Riboswitches. Science, 319(5871), 1795-1797. doi:10.1126/science.1152621