Cyclooxygenase
From Proteopedia
| Line 3: | Line 3: | ||
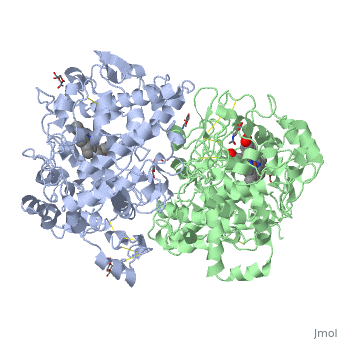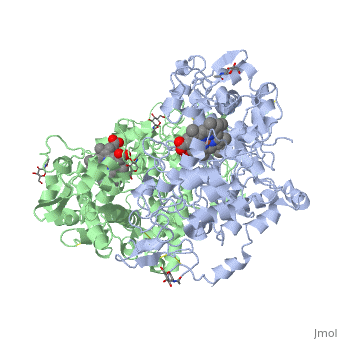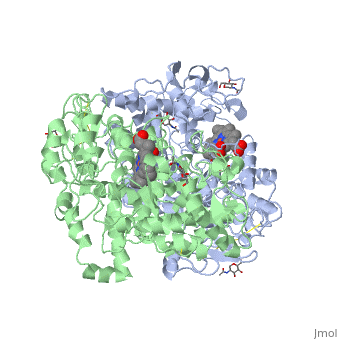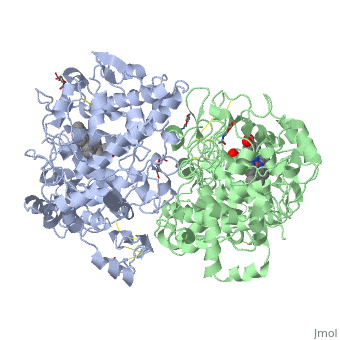
The COX monomer consists of three structural domains: the N-terminal EGF domain, a membrane binding domain (MBD) and a large C-terminal globular catalytic domain containing the heme binding site. The C-terminal segments beyond Pro583 (35 amino acids in COX-2) have not been resolved crystallographically. Collectively, these domains are made up of 25 alpha helices, seven 310 helices, four beta sheets as well as five disulfide bonds which contribute to the interface binding of the two individual monomers to complete the enzyme. | The COX monomer consists of three structural domains: the N-terminal EGF domain, a membrane binding domain (MBD) and a large C-terminal globular catalytic domain containing the heme binding site. The C-terminal segments beyond Pro583 (35 amino acids in COX-2) have not been resolved crystallographically. Collectively, these domains are made up of 25 alpha helices, seven 310 helices, four beta sheets as well as five disulfide bonds which contribute to the interface binding of the two individual monomers to complete the enzyme. | ||
| + | |||
| + | ==Protein domains== | ||
| + | ===Epidermal Growth Factor Domain=== | ||
| + | The EGF and catalytic domains create the subunit interface in the dimer and place the two MBDs in a homodimer about 25 amstrongs apart. The EGF domains create a substantial portion of the dimer interface. EGF domains are common in several families of membrane proteins and secreted proteins. Typically, the EGF domain occurs at a position in the primary sequence N-terminal to a membrane anchor, such that these domains always occur on the extracytoplasmic face of the membrane. Some authors have suggested that the EGF domains may play a role in the insertion of COX into the lipid bilayer. | ||
| + | |||
| + | ===Membrane Binding Domain=== | ||
| + | PGHS-2 associate with only one face of the membrane bilayer through a monotopic membrane binding domain (MBD) that is comprised of four short, consecutive, amphipathic α-helices (helices A–D) that include residues 59-111 in human PGHS-2 (4). Three of the four helices lie roughly in the same plane while the last helix angles “upward” into the catalytic domain. Hydrophobic and aromatic residues protrude from these helices to create a hydrophobic surface that would interact with only one face of the lipid bilayer. | ||
| + | |||
| + | ===Catalytic Domain=== | ||
| + | The catalytic domain comprises the bulk of the COX monomer and is almost entirely composed of α-helical secondary structure. As said before COX are bifunctional proteins so we can discern two types of reactions: the heme-dependent bis-oxygenase or COX reaction that converts AA to PGG2 and the subsequent peroxidase (POX) reaction that reduces the 15-hydroperoxide of PGG2 to form PGH2. | ||
| + | |||
| + | ====Peroxidase Active Site Structure==== | ||
| + | |||
| + | The <scene name='SandboxUAM/Mynewscene/4'>POX active site</scene> is in a large groove on the side opposite of the MBD. The structures of the peroxidase active sites of PGHSs are similar to those of other heme peroxidases. This site includes a heme group and the iron (III) in the center of this heme is coordinated by His-388 and by His-207. | ||
| + | |||
| + | Heme-dependent peroxidase activity is implicated in the formation of a proposed Tyr-385 radical, which is required for cyclooxygenase activity. Gln203 is also important in catalysis, although its function has not been resolved. Mutations of Gln203, His207, or His388 lead to a reduction or elimination of peroxidase activity. | ||
| + | |||
| + | The COXs bind 1 mole of ferric-protoporphyrin IX per mole monomer for full activity, as expected for a heme-dependent peroxidase. | ||
| + | |||
| + | |||
| + | ====Cyclooxygenase Active Site Structure==== | ||
| + | |||
| + | PGHS-1 and 2 monomers each contain a 25-°A hydrophobic channel that originates at the membrane binding domain and extends into the core of the globular domain. The MBD forms the mouth and the first half of the channel and allows arachidonate and O2 to enter directly from the apolar compartment of the lipid bilayer. Several amino acids composing the upper half of the channel are uniquely important in cyclooxygenase catalysis. Twenty-four residues line the hydrophobic cyclooxygenase active site with only one difference between the isozymes—Ile at position 523 in PGHS-1 and Val at position 523 in PGHS-2. Amino acids lining the hydrophobic cyclooxygenase active site channel include Leu117, Arg120, Phe205, Phe209, Val344, Ile345, Tyr348, Val349, Leu352, Ser353, Tyr355, Leu359, Phe381, Leu384, Tyr385, Trp387, Phe518, Ile/Val523, Gly526, Ala527, Ser530, Leu531, Gly533, Leu534. Only three of the channel residues are polar (Arg120, Ser353, and Ser530). Tyr 385 in its radical form is the responsible for abstracting a proton from arachidonic acid during its conversion to PGG2. Arg120, which is positioned about midway between the mouth and the apex of the active site (7), binds to the carboxylate groups of fatty acids and many NSAIDs. | ||
| + | |||
| + | |||
| + | <scene name='SandboxUAM/Mynewscene/4'>TextToBeDisplayed</scene> | ||
<Structure load='5cox' size='500' frame='true' align='right' caption='Unhibited mouse cyclooxygenase' scene='Insert optional scene name here' /> | <Structure load='5cox' size='500' frame='true' align='right' caption='Unhibited mouse cyclooxygenase' scene='Insert optional scene name here' /> | ||
Revision as of 14:42, 7 December 2010
Contents |
About this structure (1,2)
PGHSs are bifunctional homodimers. Both COX-1 and COX-2 are membrane-bound enzymes and are present on the lumenal surfaces of the endoplasmic reticulum and of the inner and outer membranes of the nuclear envelope. However, recently, it has been demonstrated in cultured endothelial cells and fibroblasts that a fraction of COX-2 protein is localized to plasma membrane in caveolae-like structures (3). The primary structure of nascent COX-2 is of 604 amino acids and then it is processed into a mature form by removal of signal peptides giving a protein of 587 amino acids. PGHS-2 is variably glycosylated at two to four sites, leading to the formation of doublets or sometimes triplets on SDS-PAGE. Murine PGHS-2 peptide is presumed to be three times at Asn56, Asn130, and Asn396(NUEVA).
The COX monomer consists of three structural domains: the N-terminal EGF domain, a membrane binding domain (MBD) and a large C-terminal globular catalytic domain containing the heme binding site. The C-terminal segments beyond Pro583 (35 amino acids in COX-2) have not been resolved crystallographically. Collectively, these domains are made up of 25 alpha helices, seven 310 helices, four beta sheets as well as five disulfide bonds which contribute to the interface binding of the two individual monomers to complete the enzyme.
Protein domains
Epidermal Growth Factor Domain
The EGF and catalytic domains create the subunit interface in the dimer and place the two MBDs in a homodimer about 25 amstrongs apart. The EGF domains create a substantial portion of the dimer interface. EGF domains are common in several families of membrane proteins and secreted proteins. Typically, the EGF domain occurs at a position in the primary sequence N-terminal to a membrane anchor, such that these domains always occur on the extracytoplasmic face of the membrane. Some authors have suggested that the EGF domains may play a role in the insertion of COX into the lipid bilayer.
Membrane Binding Domain
PGHS-2 associate with only one face of the membrane bilayer through a monotopic membrane binding domain (MBD) that is comprised of four short, consecutive, amphipathic α-helices (helices A–D) that include residues 59-111 in human PGHS-2 (4). Three of the four helices lie roughly in the same plane while the last helix angles “upward” into the catalytic domain. Hydrophobic and aromatic residues protrude from these helices to create a hydrophobic surface that would interact with only one face of the lipid bilayer.
Catalytic Domain
The catalytic domain comprises the bulk of the COX monomer and is almost entirely composed of α-helical secondary structure. As said before COX are bifunctional proteins so we can discern two types of reactions: the heme-dependent bis-oxygenase or COX reaction that converts AA to PGG2 and the subsequent peroxidase (POX) reaction that reduces the 15-hydroperoxide of PGG2 to form PGH2.
Peroxidase Active Site Structure
The is in a large groove on the side opposite of the MBD. The structures of the peroxidase active sites of PGHSs are similar to those of other heme peroxidases. This site includes a heme group and the iron (III) in the center of this heme is coordinated by His-388 and by His-207.
Heme-dependent peroxidase activity is implicated in the formation of a proposed Tyr-385 radical, which is required for cyclooxygenase activity. Gln203 is also important in catalysis, although its function has not been resolved. Mutations of Gln203, His207, or His388 lead to a reduction or elimination of peroxidase activity.
The COXs bind 1 mole of ferric-protoporphyrin IX per mole monomer for full activity, as expected for a heme-dependent peroxidase.
Cyclooxygenase Active Site Structure
PGHS-1 and 2 monomers each contain a 25-°A hydrophobic channel that originates at the membrane binding domain and extends into the core of the globular domain. The MBD forms the mouth and the first half of the channel and allows arachidonate and O2 to enter directly from the apolar compartment of the lipid bilayer. Several amino acids composing the upper half of the channel are uniquely important in cyclooxygenase catalysis. Twenty-four residues line the hydrophobic cyclooxygenase active site with only one difference between the isozymes—Ile at position 523 in PGHS-1 and Val at position 523 in PGHS-2. Amino acids lining the hydrophobic cyclooxygenase active site channel include Leu117, Arg120, Phe205, Phe209, Val344, Ile345, Tyr348, Val349, Leu352, Ser353, Tyr355, Leu359, Phe381, Leu384, Tyr385, Trp387, Phe518, Ile/Val523, Gly526, Ala527, Ser530, Leu531, Gly533, Leu534. Only three of the channel residues are polar (Arg120, Ser353, and Ser530). Tyr 385 in its radical form is the responsible for abstracting a proton from arachidonic acid during its conversion to PGG2. Arg120, which is positioned about midway between the mouth and the apex of the active site (7), binds to the carboxylate groups of fatty acids and many NSAIDs.
|
NSAIDs
| Drug | Coefficient of selectivity (IC50Cox-1/IC50Cox-2) |
|---|---|
| Ketorolac | |
| Naproxen | |
| Ibuprofen | |
| Indometacin | |
| Acetylsalicylic acid | |
| Diclofenac | |
| Valdecoxib | |
| Etoricoxib | |
| Pharmacologic group | Drug |
|---|---|
| Salicylates | Acetylsalicylic acid |
| Propionic | Naproxen |
| Ibuprofen | |
| Para-aminophenols | Paracetamol |
| Indolacetic | Indometacin |
| Pirrolacetic | Ketorolac |
| Phenilacetic | Diclofenac |
| Piranoidacetic | Etodolac |
| Anthranilic | Mefenamic acid |
| Nicotinic | Clonixin |
| Sulfonanilides | Nimesulide |
Reference
- Ghosh N, Chaki R, Mandal V, Mandal SC. COX-2 as a target for cancer chemotherapy. Pharmacol Rep. 2010 Mar-Apr;62(2):233-44. PMID:20508278
- Smith WL, DeWitt DL, Garavito RM. Cyclooxygenases: structural, cellular, and molecular biology. Annu Rev Biochem. 2000;69:145-82. PMID:10966456 doi:10.1146/annurev.biochem.69.1.145
- Rang HP, Dale MM, Ritter JM, Flower RJ. 2008. Pharmacology. Elsevier. 6th edition. 844 p.
- Smith WL, Langenbach R. Why there are two cyclooxygenase isozymes. J Clin Invest. 2001 Jun;107(12):1491-5. PMID:11413152 doi:10.1172/JCI13271
- Chandrasekharan NV, Dai H, Roos KL, Evanson NK, Tomsik J, Elton TS, Simmons DL. COX-3, a cyclooxygenase-1 variant inhibited by acetaminophen and other analgesic/antipyretic drugs: cloning, structure, and expression. Proc Natl Acad Sci U S A. 2002 Oct 15;99(21):13926-31. Epub 2002 Sep 19. PMID:12242329 doi:10.1073/pnas.162468699
- Garavito RM, Mulichak AM. The structure of mammalian cyclooxygenases. Annu Rev Biophys Biomol Struct. 2003;32:183-206. Epub 2003 Feb 5. PMID:12574066 doi:10.1146/annurev.biophys.32.110601.141906
- Perrone G, Zagami M, Altomare V, Battista C, Morini S, Rabitti C. COX-2 localization within plasma membrane caveolae-like structures in human lobular intraepithelial neoplasia of the breast. Virchows Arch. 2007 Dec;451(6):1039-45. Epub 2007 Sep 13. PMID:17851687 doi:10.1007/s00428-007-0506-4
- Spencer AG, Thuresson E, Otto JC, Song I, Smith T, DeWitt DL, Garavito RM, Smith WL. The membrane binding domains of prostaglandin endoperoxide H synthases 1 and 2. Peptide mapping and mutational analysis. J Biol Chem. 1999 Nov 12;274(46):32936-42. PMID:10551860
- Luong C, Miller A, Barnett J, Chow J, Ramesha C, Browner MF. Flexibility of the NSAID binding site in the structure of human cyclooxygenase-2. Nat Struct Biol. 1996 Nov;3(11):927-33. PMID:8901870
- Kurumbail RG, Stevens AM, Gierse JK, McDonald JJ, Stegeman RA, Pak JY, Gildehaus D, Miyashiro JM, Penning TD, Seibert K, Isakson PC, Stallings WC. Structural basis for selective inhibition of cyclooxygenase-2 by anti-inflammatory agents. Nature. 1996 Dec 19-26;384(6610):644-8. PMID:8967954 doi:http://dx.doi.org/10.1038/384644a0
Proteopedia Page Contributors and Editors (what is this?)
Cristina Murga, Michal Harel, David Canner, María Laura Saiz Álvarez, Alexander Berchansky