Gyrase
From Proteopedia
| Line 50: | Line 50: | ||
==References== | ==References== | ||
<References /> | <References /> | ||
| + | |||
| + | [[Category:Topic Page]] | ||
Revision as of 07:10, 23 December 2010
|
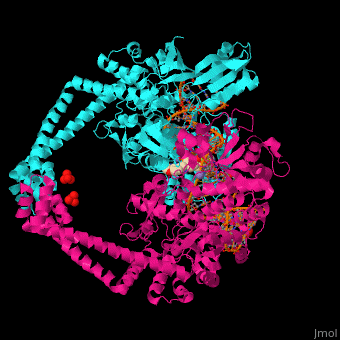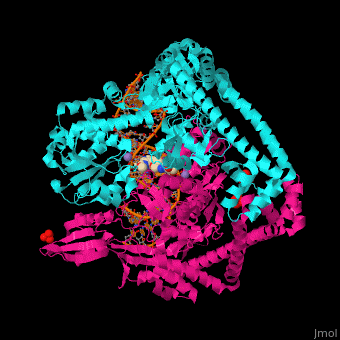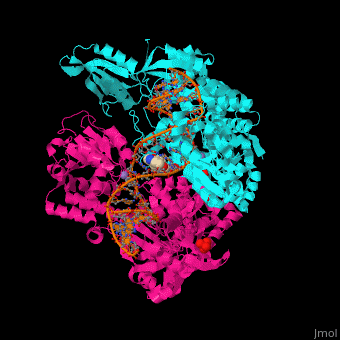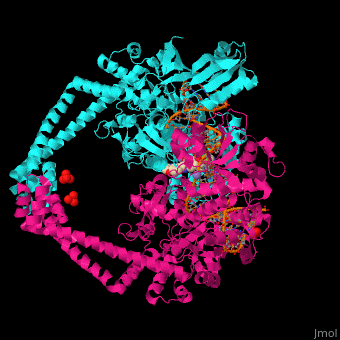
Gyrase (Gyr) is a type of topoisomerase II in prokaryotes which unwinds double stranded DNA. The DNA Gyr cutting allows the formation of a negative DNA supercoil which enables replication of DNA. Gyr consists of 2 subunits: GyrA and GyrB. Reverse gyrase (Top-RG) is a type of topoisomerase I which catalyses the formation of positive DNA supercoil. [1]
Contents |
3D Structure of Gyrase
Gyrase Subunit A
3l6v – GyrA C-terminal – Xanthomonas campestris
2wl2 – EcGyrA N-terminal+simocylinone – Escherichia coli
1ajb - EcGyrA N-terminal+novobiocin
1zi0, 1ab4 - EcGyrA C-terminal
3ku8 – EcGyrA fragment+CcdB
1x75 – EcGyrA14+CcdB
3kua - GyrA fragment+CcdB – Vibrio fischeri
3ilw, 3ifz - MtGyrA N-terminal – Mycobacterium tuberculosis
1suu - GyrA C-terminal – Borrelia burgdorferi
3no0 - GyrA C-terminal – Aquifex aeolicus
Gyrase Subunit B
3g75, 3g7b, 3g7e – GyrB+thiazole inhibitor – Staphylococcus aureus
2zjt, 3ig0, 3m4i - MtGyrB C-terminal
3cwv – GyrB truncated – Myxococcus xanthus
1kzn, 1ei1 - EcGyrB N-terminal+clorobiocin
1aj6 - EcGyrB N-terminal+novobiocin
1kij – GyrB domain+novobiocin – Thermus thermophilus
Gyrase Subunit A+Subunit B
2xco, 2xcq - SaGyrB C-terminal-SaGyrA N-terminal fusion
2xcr, 2xcs - SaGyrB C-terminal-SaGyrA N-terminal fusion (mutant)+DNA
2xct - SaGyrB C-terminal-SaGyrA N-terminal fusion (mutant) +DNA+ ciprofloxacin
3nuh – EcGyrA+EcGyrB
Reverse Gyrase
1gku – AfTop-RG – Archaeoglobus fulgidus
1gl9 - AfTop-RG+ADPNP
3oiy – Top-RG helicase domain – Thermotoga maritima
Additional Resources
For additional information, see: Bacterial Infections
References
- ↑ Gore J, Bryant Z, Stone MD, Nollmann M, Cozzarelli NR, Bustamante C. Mechanochemical analysis of DNA gyrase using rotor bead tracking. Nature. 2006 Jan 5;439(7072):100-4. PMID:16397501 doi:10.1038/nature04319
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, David Canner, Joel L. Sussman