We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Potassium Channel
From Proteopedia
(Difference between revisions)
| Line 4: | Line 4: | ||
Potassium channels possess two traits that are seemingly mutually exclusive. Firstly, potassium channels have exquisite selectivity, with an amazing 10,000 fold selectivity for K<sup>+</sup> ions over sodium ions. Considering the only difference by which potassium ions can be differentiated from sodium ions is potassium ions’ 1.33Å Pauling radius vs. Sodium’s .95Å radius, the selectivity of potassium channels is remarkable.<ref name="Doyle"/> Second, despite its remarkable selectivity, potassium channels allow for the transfer of K<sup>+</sup> ions across the cell membrane at a rate of nearly 10<sup>8</sup> per second, nearly at the diffusion rate limit.<ref name="Long">PMID: 18004376</ref> Potassium channels are able to achieve these remarkable feats due to its amazing structural architecture contains several remarkable features which not only can sense the voltage potential across a membrane, but also selectively ferry K<sup>+</sup> ions without any outside energy expenditure. | Potassium channels possess two traits that are seemingly mutually exclusive. Firstly, potassium channels have exquisite selectivity, with an amazing 10,000 fold selectivity for K<sup>+</sup> ions over sodium ions. Considering the only difference by which potassium ions can be differentiated from sodium ions is potassium ions’ 1.33Å Pauling radius vs. Sodium’s .95Å radius, the selectivity of potassium channels is remarkable.<ref name="Doyle"/> Second, despite its remarkable selectivity, potassium channels allow for the transfer of K<sup>+</sup> ions across the cell membrane at a rate of nearly 10<sup>8</sup> per second, nearly at the diffusion rate limit.<ref name="Long">PMID: 18004376</ref> Potassium channels are able to achieve these remarkable feats due to its amazing structural architecture contains several remarkable features which not only can sense the voltage potential across a membrane, but also selectively ferry K<sup>+</sup> ions without any outside energy expenditure. | ||
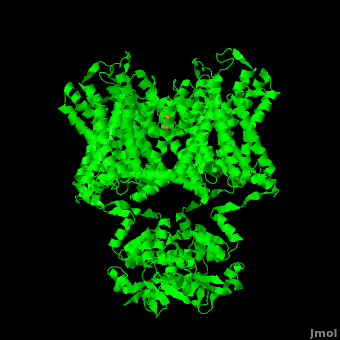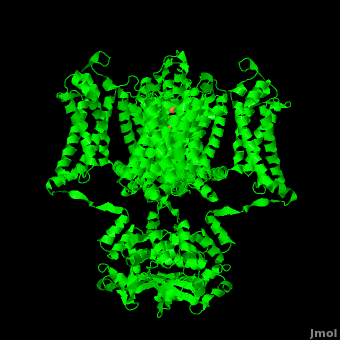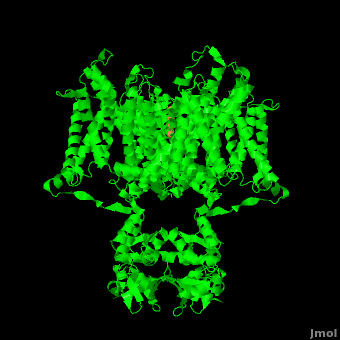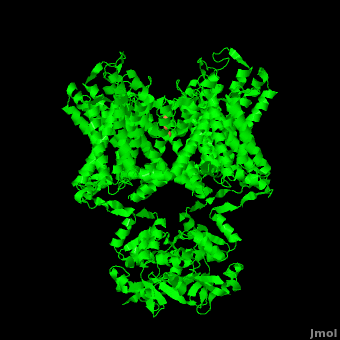
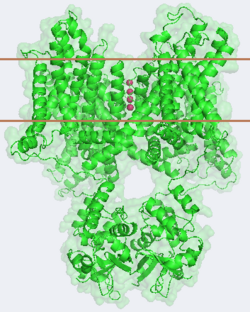
| - | The overall structure of the voltage gated potassium channel can be seen in the image at the left. It contains several key features which will be analyzed. Primarily, a <scene name='Potassium_Channel/Trans/ | + | The overall structure of the voltage gated potassium channel can be seen in the image at the left. It is comprised of 4 identical subunits and contains several key features which will be analyzed. Primarily, a <scene name='Potassium_Channel/Trans/3'>transmembrane region</scene> marked between the parallel lines in the figure. This region houses the <scene name='Potassium_Channel/Pore_opening/5'>channel pore</scene>, composed of interwoven helices in a teepee conformation, the all-important <scene name='Potassium_Channel/Selectivity_filter_opening/2'>“selectivity filter”</scene>, providing the channel with its remarkable 10,00 fold selectivity for K<sup>+</sup> ions over Na<sup>+</sup> ions and the <scene name='Potassium_Channel/Voltage_sensors_opening/4'>“voltage sensor”</scene> which is uses well placed arginine and acidic residues to determine the membrane polarity and open/close the channel in response.<ref name="Long"/> |
====Selectivity Filter and Pore==== | ====Selectivity Filter and Pore==== | ||
| - | It is instructive to follow the path of a potassium ion as it enters the cell through the <scene name='Potassium_Channel/Potassium_out/ | + | It is instructive to follow the path of a potassium ion as it enters the cell through the <scene name='Potassium_Channel/Potassium_out/3'>potassium channel</scene>. Upon <scene name='Potassium_Channel/Into_pore/3'>entering the channel</scene>, the K<sup>+</sup> ion first comes into contact with the <scene name='Potassium_Channel/From_extra/2'>selectivity filter</scene>. The solved structure of the potassium channel by MacKinnon et al. revealed where the channels remarkable selectivity comes from. When entering the selectivity filter, K<sup>+</sup> ions are first dehydrated, shedding up to 8 waters. To stabilize these naked ions, **a number of carbonyl oxygens** bind the K<sup>+</sup> ions. The **distance between** K<sup>+</sup> ion and carbonyl oxygen is at the perfect width to accommodate K<sup>+</sup> ions but not Na<sup>+</sup> ions which are too small. If a Na<sup>+</sup> ion were to lose it’s water shell, the carbonyl oxygens could not successfully stabilize it in its naked form and thus it is energetically unfavorable for a Na<sup>+</sup> ion to enter the channel. There is room within the selectivity filter for four K<sup>+</sup> ions. This, as it turns out, is crucial as the presence of the positive cations in close proximity to one another effectively pushes the potassium ions through the filter via electrostatic forces. This helps explain how the potassium channel can have such a rapid turnover rate.<ref name="Doyle"/> Also, the **natural polarity of the helices**, with the **carbonyl oxygens pointing down the pore**, helps drag the potassium ions through the channel quickly. When exposed to a low concentration of potassium, the channel assumes a **“low concentration” conformation** (LOW CONFORMATION STRUCCTURE) which is sealed shut.<ref name="Zhou"/> |
The selectivity filter only makes up only the beginning of the **channel pore**. With the exception of the selectivity filter, the pore lining is **mainly hydrophobic**. This hydrophobic lining provides an inert surface over which the diffusing ion can slide unimpaired. Immediately following the selectivity filter is an **aqueous cavity**. K<sup>+</sup> ions, after passing through the filter, rehydrate in this cavity, helping overcome much of the energetic difficulty of having a positively charged cation within a hydrophobic membrane. At the bottom of the 34Å pore containing transmembrane region lies a number of **aromatic residues** which help form a seal between the pore and the intracellular cytoplasm.<ref name="Doyle"/> | The selectivity filter only makes up only the beginning of the **channel pore**. With the exception of the selectivity filter, the pore lining is **mainly hydrophobic**. This hydrophobic lining provides an inert surface over which the diffusing ion can slide unimpaired. Immediately following the selectivity filter is an **aqueous cavity**. K<sup>+</sup> ions, after passing through the filter, rehydrate in this cavity, helping overcome much of the energetic difficulty of having a positively charged cation within a hydrophobic membrane. At the bottom of the 34Å pore containing transmembrane region lies a number of **aromatic residues** which help form a seal between the pore and the intracellular cytoplasm.<ref name="Doyle"/> | ||
Revision as of 20:36, 7 March 2011
| |||||||||||
Additional Structures of Potassium Channels
For Additional Structures, See: Potassium Channels
Additional Resources
For Additional Information, See: Membrane Channels & Pumps
References
- ↑ 1.0 1.1 Zhou Y, Morais-Cabral JH, Kaufman A, MacKinnon R. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution. Nature. 2001 Nov 1;414(6859):43-8. PMID:11689936 doi:http://dx.doi.org/10.1038/35102009
- ↑ 2.0 2.1 2.2 2.3 Doyle DA, Morais Cabral J, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998 Apr 3;280(5360):69-77. PMID:9525859
- ↑ Jiang Y, Lee A, Chen J, Ruta V, Cadene M, Chait BT, MacKinnon R. X-ray structure of a voltage-dependent K+ channel. Nature. 2003 May 1;423(6935):33-41. PMID:12721618 doi:http://dx.doi.org/10.1038/nature01580
- ↑ Waters MF, Minassian NA, Stevanin G, Figueroa KP, Bannister JP, Nolte D, Mock AF, Evidente VG, Fee DB, Muller U, Durr A, Brice A, Papazian DM, Pulst SM. Mutations in voltage-gated potassium channel KCNC3 cause degenerative and developmental central nervous system phenotypes. Nat Genet. 2006 Apr;38(4):447-51. Epub 2006 Feb 26. PMID:16501573 doi:ng1758
- ↑ 5.0 5.1 5.2 5.3 Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007 Nov 15;450(7168):376-82. PMID:18004376 doi:http://dx.doi.org/10.1038/nature06265
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, David Canner, Joel L. Sussman, Alexander Berchansky, Ann Taylor