Ouabain
From Proteopedia
| Line 2: | Line 2: | ||
{{Template:Oberholser_Sandbox_Reservation}} | {{Template:Oberholser_Sandbox_Reservation}} | ||
<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | <!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | ||
| - | + | ||
=Ouabain= | =Ouabain= | ||
| - | <scene name='Sandbox_60/Ouabain_3d/1'>Ouabain</scene> is a cardiac glycoside that inhibits | + | <scene name='Sandbox_60/Ouabain_3d/1'>Ouabain</scene> is a cardiac glycoside that inhibits ATP-dependent sodium-potassium exchange across cell membranes. The binding of ouabain to the sodium-potassium pump (also called Na+/K+ ATPase) prevents the conformational changes necessary for its proper function. This affects intracellular ion composition in various ways with various effects, depending on the cell and the dosage. The compound has therefore been utilized in medicine, both as a drug and in research related to active membrane transport. |
| - | [[Image: | + | |
| + | <Structure load='3n23' size='500' frame='true' align='right' caption='Insert caption here' scene='Insert optional scene name here' /> | ||
| + | |||
| + | ==Structure and Na+/K+ Pump Binding== | ||
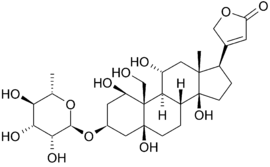
| + | Below is the structure of oubain in two dimensions. The molecule consists of a sugar bound to a modified cholesterol by a glycosidic linkage (hence ''glycoside''). The hydroxyl groups surrounding much of the molecule, along with the esters at either end, contribute to its binding to the membrane bound sodium-potassium pump. <scene name='Sandbox_60/Ouabain_3d/1'>Ouabain</scene> can also be seen in three dimensions. With the molecular geometry and stereochemistry displayed in this way, one can see more clearly the distribution of polar carbon-oxygen and non-polar carbon-carbon bonds in the space surrounding the molecule. This makes visualizing the binding of the inhibitor much easier.[[Image:Ouabain.png|thumb|left|upright=1.5]] Ouabain is <scene name='Sandbox_60/Drug_in_complex/1'>bound</scene> to the protein along the inside of an alpha-helix bundle. <scene name='Sandbox_60/Drug_in_complex_np/1'>Non-polar</scene> components of residues help somewhat in coordinating ouabain through Van der Waals forces, but <scene name='Sandbox_60/Drug_in_complex_p/1'>polar</scene> residues, glutamine, aspartic acid, and threonine, along with the amide bond of an alanine, surround the hydroxyl and carbonyl groups of the ligand, forming hydrogen bonds of 2 to 4 angstroms in length. This not only holds the drug in place, but prevents conformational change necessary for the function of the protein. | ||
| + | |||
| + | ==Na+/K+ ATPase and Cardiac Muscle Contraction== | ||
| + | Na+/K+ ATPase actively transports potassium into the cell and sodium to the extra-cellular space according to the general scheme depicted below. This transmembrane protein is composed of two subunits, alpha and beta, that may function as a tetramer ''in vivo''. Here, the ''in vitro'' crystal structure shows the alpha chain in pink/blue and the beta chain in yellow/green. The alpha subunit plays the leading role in the functioning of the protein. Inside the transmembrane domain, residues coordinate sodium or potassium ions. The large domain on the cytoplasmic side of the bilayer (furthest from its own beta subunit) is responsible for binding and hydrolyzing ATP with the aid of a magnesium cofactor. The energy released from this cleavage is transfered via conformational changes to the transmembrane domain, where ions are forced against their concentration gradient. On the extracellular side, the beta subunit moves to prevent the dissociation of potassium ions during their entry into the cell. A third gamma subunit (purple/orange), along with the beta subunit, contributes to the anchoring of transmembrane domains to the phospholipid bilayer. | ||
| + | Normal functioning Na+/K+ ATPase changes unidirectionally between two phases, E1 and E2. In the E1 phase, it binds Na+ and ATP on the inside of the cell. After hydrolysis, the phosphorelated protein ejects ADP and changes conformation to E2, allowing for 3Na+ to dissociate on the extracellular side of the membrane. The protein then binds 2K+ and hydrolyzes the bound inorganic phosphate, causing the reversion to E1. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | [[Image:498px-Scheme sodium-potassium pump-en-2.svg.png|thumb|right|upright=1.5]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <scene name='Sandbox_60/Drug_in_complex_situated/1'>TextToBeDisplayed</scene> | ||
Revision as of 22:01, 10 March 2011
| Please do NOT make changes to this Sandbox. Sandboxes 30-60 are reserved for use by Biochemistry 410 & 412 at Messiah College taught by Dr. Hannah Tims during Fall 2012 and Spring 2013. |
Ouabain
is a cardiac glycoside that inhibits ATP-dependent sodium-potassium exchange across cell membranes. The binding of ouabain to the sodium-potassium pump (also called Na+/K+ ATPase) prevents the conformational changes necessary for its proper function. This affects intracellular ion composition in various ways with various effects, depending on the cell and the dosage. The compound has therefore been utilized in medicine, both as a drug and in research related to active membrane transport.
|
Structure and Na+/K+ Pump Binding
Below is the structure of oubain in two dimensions. The molecule consists of a sugar bound to a modified cholesterol by a glycosidic linkage (hence glycoside). The hydroxyl groups surrounding much of the molecule, along with the esters at either end, contribute to its binding to the membrane bound sodium-potassium pump. can also be seen in three dimensions. With the molecular geometry and stereochemistry displayed in this way, one can see more clearly the distribution of polar carbon-oxygen and non-polar carbon-carbon bonds in the space surrounding the molecule. This makes visualizing the binding of the inhibitor much easier. Ouabain is to the protein along the inside of an alpha-helix bundle. components of residues help somewhat in coordinating ouabain through Van der Waals forces, but residues, glutamine, aspartic acid, and threonine, along with the amide bond of an alanine, surround the hydroxyl and carbonyl groups of the ligand, forming hydrogen bonds of 2 to 4 angstroms in length. This not only holds the drug in place, but prevents conformational change necessary for the function of the protein.Na+/K+ ATPase and Cardiac Muscle Contraction
Na+/K+ ATPase actively transports potassium into the cell and sodium to the extra-cellular space according to the general scheme depicted below. This transmembrane protein is composed of two subunits, alpha and beta, that may function as a tetramer in vivo. Here, the in vitro crystal structure shows the alpha chain in pink/blue and the beta chain in yellow/green. The alpha subunit plays the leading role in the functioning of the protein. Inside the transmembrane domain, residues coordinate sodium or potassium ions. The large domain on the cytoplasmic side of the bilayer (furthest from its own beta subunit) is responsible for binding and hydrolyzing ATP with the aid of a magnesium cofactor. The energy released from this cleavage is transfered via conformational changes to the transmembrane domain, where ions are forced against their concentration gradient. On the extracellular side, the beta subunit moves to prevent the dissociation of potassium ions during their entry into the cell. A third gamma subunit (purple/orange), along with the beta subunit, contributes to the anchoring of transmembrane domains to the phospholipid bilayer. Normal functioning Na+/K+ ATPase changes unidirectionally between two phases, E1 and E2. In the E1 phase, it binds Na+ and ATP on the inside of the cell. After hydrolysis, the phosphorelated protein ejects ADP and changes conformation to E2, allowing for 3Na+ to dissociate on the extracellular side of the membrane. The protein then binds 2K+ and hydrolyzes the bound inorganic phosphate, causing the reversion to E1.