We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Journal:PLoS ONE:1
From Proteopedia
(Difference between revisions)

| Line 7: | Line 7: | ||
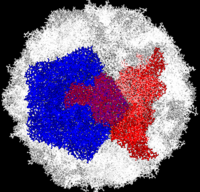
[[Image:Polio.png|left|200px|thumb|Complete Poliovirus 2 Viron based on PDB entry [[1eah]], <font color='red'><b>example of 3-fold symmetry is in red</b></font>, <font color='blue'><b>example of 5-fold symmetry is in blue</b></font> ]] | [[Image:Polio.png|left|200px|thumb|Complete Poliovirus 2 Viron based on PDB entry [[1eah]], <font color='red'><b>example of 3-fold symmetry is in red</b></font>, <font color='blue'><b>example of 5-fold symmetry is in blue</b></font> ]] | ||
| - | Poliovirus is a member of the ''Picornaviridae''. Like other members of the ''Picornaviridae'', poliovirus RNA is encapsulated in an icosahedral structure with axes of <scene name='Journal:PLoS_ONE:1/F43/ | + | Poliovirus is a member of the ''Picornaviridae''. Like other members of the ''Picornaviridae'', poliovirus RNA is encapsulated in an icosahedral structure with axes of <scene name='Journal:PLoS_ONE:1/F43/2'>three-fold</scene> and <scene name='Journal:PLoS_ONE:1/F45/1'>five-fold</scene> symmetry formed from 60 <scene name='Journal:PLoS_ONE:1/F1/3'>capsomeres containing one copy</scene> each of viral capsid proteins VP1 (light blue), VP2 (pale green), VP3 (light orange) and VP4(light magenta). The binding site for the human poliovirus receptor is located in a canyon at the five-fold axis of symmetry. The VP1 of picornaviruses contain a hydrophobic pocket that is accessed through this canyon. This pocket is normally occupied by pocket factors, sphingosine-like molecules including |
palmitic and myristic acids and hydrophobic compounds, that stabilize the capsid and whose removal is a necessary prerequisite for uncoating. | palmitic and myristic acids and hydrophobic compounds, that stabilize the capsid and whose removal is a necessary prerequisite for uncoating. | ||
Revision as of 15:03, 27 March 2011
| |||||||||||
- ↑ DOI
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.