This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Journal:PLoS ONE:1
From Proteopedia
(Difference between revisions)

| Line 7: | Line 7: | ||
<hr/> | <hr/> | ||
<b>Molecular Tour</b><br> | <b>Molecular Tour</b><br> | ||
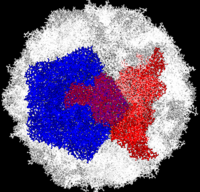
| - | Poliovirus is a member of the ''Picornaviridae''. Like other members of the ''Picornaviridae'', poliovirus RNA is encapsulated in an icosahedral structure with axes of <scene name='Journal:PLoS_ONE:1/F43/2'>three-fold</scene> and <scene name='Journal:PLoS_ONE:1/F45/1'>five-fold</scene> symmetry formed from 60 <scene name='Journal:PLoS_ONE:1/F1/3'>capsomeres containing one copy</scene> each of viral capsid proteins VP1 (light blue), VP2 (pale green), VP3 (light orange) and VP4(light magenta). The binding site for the human poliovirus receptor is located <scene name='Journal:PLoS_ONE:1/F45/2'>in a canyon at the five-fold axis of symmetry</scene>. The VP1 of picornaviruses contain a hydrophobic pocket that is accessed through this canyon. This pocket is normally occupied by <scene name='Journal:PLoS_ONE:1/F45/3'>pocket factors, sphingosine-like molecules including palmitic and myristic acids and hydrophobic compounds, that stabilize the capsid and whose removal is a necessary prerequisite for uncoating</scene>. The <scene name='Journal:PLoS_ONE:1/F45/4'>broad-spectrum antiviral agent SCH48973</scene> is observed binding in a pocket within the beta-barrel of VP1 ([[1eah]]), in approximately the same location that natural pocket factors bind to polioviruses. SCH48973 forms predominantly hydrophobic interactions with the pocket residues. Flavanoids and flavonoids were shown to have antiviral activity ''in vitro'' against several picornaviruses including type 2 polioviruses. These compounds act primarily by occupying the hydrophobic pocket, thus interfering with virus uncoating. Compounds <scene name='Journal:PLoS_ONE:1/Iso/2'>3(2H)-Isoflavene (C2)</scene> and 6-chloro-3(2H)-Isoflavene (C10) are relatively similar to <scene name='Journal:PLoS_ONE:1/Sc/1'>SCH48973</scene> and exert their antiviral effect by interacting with amino acids in the viral capsid pockets. Ambiguous vaccine-derived poliovirus 2 (aVDPV) isolate <scene name='Journal:PLoS_ONE:1/F1/1'>SD-06-10</scene> was resistant to the antiviral effects of both C2 and C10 compounds. The ''in silico'' translated amino acid sequences of the capsid proteins of all of the aVDPV2 isolates and Sabin 2 (AY184220) were compared and amino acid substitutions in capsid proteins that were unique to SD-06-10 were determined. The positions of these unique substitutions were mapped onto the coordinates of type 2 poliovirus ([[1eah]]) using MacPyMol and <scene name='Journal:PLoS_ONE:1/F1/4'>are represented by red spheres</scene>. <font color='blue'><b>The hydrophobic pocket is represented by blue spheres</b></font> and the position of amino-acid substitutions that were previously shown to result in <font color='yellow'><b>loss of sensitivity</b></font> to or <font color=' | + | Poliovirus is a member of the ''Picornaviridae''. Like other members of the ''Picornaviridae'', poliovirus RNA is encapsulated in an icosahedral structure with axes of <scene name='Journal:PLoS_ONE:1/F43/2'>three-fold</scene> and <scene name='Journal:PLoS_ONE:1/F45/1'>five-fold</scene> symmetry formed from 60 <scene name='Journal:PLoS_ONE:1/F1/3'>capsomeres containing one copy</scene> each of viral capsid proteins VP1 (light blue), VP2 (pale green), VP3 (light orange) and VP4(light magenta). The binding site for the human poliovirus receptor is located <scene name='Journal:PLoS_ONE:1/F45/2'>in a canyon at the five-fold axis of symmetry</scene>. The VP1 of picornaviruses contain a hydrophobic pocket that is accessed through this canyon. This pocket is normally occupied by <scene name='Journal:PLoS_ONE:1/F45/3'>pocket factors, sphingosine-like molecules including palmitic and myristic acids and hydrophobic compounds, that stabilize the capsid and whose removal is a necessary prerequisite for uncoating</scene>. The <scene name='Journal:PLoS_ONE:1/F45/4'>broad-spectrum antiviral agent SCH48973</scene> is observed binding in a pocket within the beta-barrel of VP1 ([[1eah]]), in approximately the same location that natural pocket factors bind to polioviruses. SCH48973 forms predominantly hydrophobic interactions with the pocket residues. Flavanoids and flavonoids were shown to have antiviral activity ''in vitro'' against several picornaviruses including type 2 polioviruses. These compounds act primarily by occupying the hydrophobic pocket, thus interfering with virus uncoating. Compounds <scene name='Journal:PLoS_ONE:1/Iso/2'>3(2H)-Isoflavene (C2)</scene> and 6-chloro-3(2H)-Isoflavene (C10) are relatively similar to <scene name='Journal:PLoS_ONE:1/Sc/1'>SCH48973</scene> and exert their antiviral effect by interacting with amino acids in the viral capsid pockets. Ambiguous vaccine-derived poliovirus 2 (aVDPV) isolate <scene name='Journal:PLoS_ONE:1/F1/1'>SD-06-10</scene> was resistant to the antiviral effects of both C2 and C10 compounds. The ''in silico'' translated amino acid sequences of the capsid proteins of all of the aVDPV2 isolates and Sabin 2 (AY184220) were compared and amino acid substitutions in capsid proteins that were unique to SD-06-10 were determined. The positions of these unique substitutions were mapped onto the coordinates of type 2 poliovirus ([[1eah]]) using MacPyMol and <scene name='Journal:PLoS_ONE:1/F1/4'>are represented by red spheres</scene>. <font color='blue'><b>The hydrophobic pocket is represented by blue spheres</b></font> and the position of amino-acid substitutions that were previously shown to result in <font color='yellow'><b>loss of sensitivity</b></font> to or <font color='lime'><b>dependence</b></font> on isoflavenes are indicated by the yellow and green spheres, respectively. |
<scene name='Journal:PLoS_ONE:1/F41/2'>TextToBeDisplayed</scene> | <scene name='Journal:PLoS_ONE:1/F41/2'>TextToBeDisplayed</scene> | ||
Revision as of 18:44, 27 March 2011
Complete Poliovirus 2 Viron (capsid) based on PDB entry 1eah, example of 3-fold symmetry is in red, example of 5-fold symmetry is in blue.
| |||||||||||
- ↑ DOI
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.