Copper Amine Oxidase
From Proteopedia
| Line 4: | Line 4: | ||
| - | 2d1w is a [http://en.wikipedia.org/wiki/Amine_oxidase_%28copper-containing%29 copper amine oxidase] found in [http://en.wikipedia.org/wiki/Arthrobacter_globiformis Arthrobacter globiformis]. | ||
== Structure == | == Structure == | ||
{{STRUCTURE_2d1w | PDB=2d1w | SCENE= }} | {{STRUCTURE_2d1w | PDB=2d1w | SCENE= }} | ||
| + | 2d1w is a [http://en.wikipedia.org/wiki/Amine_oxidase_%28copper-containing%29 copper amine oxidase] found in [http://en.wikipedia.org/wiki/Arthrobacter_globiformis Arthrobacter globiformis]. | ||
| + | The structure of this enzyme was determined by Murakawa et al. in 2005 <ref>PMID:16487484</ref>. It consists of a dimer containing 638 residues, and there is a copper ligand located near the center of each subunit. | ||
=== Ligand === | === Ligand === | ||
| - | + | The Cu<sup>2+</sup> ligand<scene name='Sandbox_Reserved_331/Copper_ligand/5'> (shown here)</scene> is coordinated by three histidine residues and is located near the active site of the enzyme. | |
| - | + | ||
=== Modified Residue === | === Modified Residue === | ||
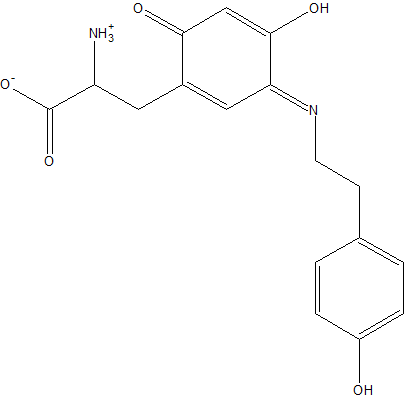
| + | Residue 382 consists of a modified residue, derived from tyrosine, that joins two peptide chains, much like a disulfide bridge would. | ||
[[Image:TTS.png|left|frame|alt=3-((3E)-4-HYDROXY-3-{[2-(4-HYDROXYPHENYL)ETHYL]IMINO}-6-OXOCYCLOHEXA-1,4-DIEN-1-YL)ALANINE.|Residue 382 is a modified tyrosine residue.]] | [[Image:TTS.png|left|frame|alt=3-((3E)-4-HYDROXY-3-{[2-(4-HYDROXYPHENYL)ETHYL]IMINO}-6-OXOCYCLOHEXA-1,4-DIEN-1-YL)ALANINE.|Residue 382 is a modified tyrosine residue.]] | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
Revision as of 23:39, 29 March 2011
| This Sandbox is Reserved from January 10, 2010, through April 10, 2011 for use in BCMB 307-Proteins course taught by Andrea Gorrell at the University of Northern British Columbia, Prince George, BC, Canada. |
To get started:
More help: Help:Editing |
Contents |
Structure
Template:STRUCTURE 2d1w 2d1w is a copper amine oxidase found in Arthrobacter globiformis. The structure of this enzyme was determined by Murakawa et al. in 2005 [1]. It consists of a dimer containing 638 residues, and there is a copper ligand located near the center of each subunit.
Ligand
The Cu2+ ligand is coordinated by three histidine residues and is located near the active site of the enzyme.
Modified Residue
Residue 382 consists of a modified residue, derived from tyrosine, that joins two peptide chains, much like a disulfide bridge would.
Reaction
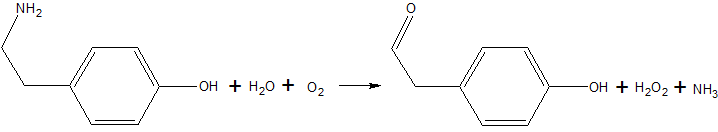
Copper amine oxidase catalyzes the oxidation of a primary amine to the corresponding aldehyde, yielding hydrogen peroxide and free ammonia. An example of this is the oxidation of tyramine:
References
- ↑ Murakawa T, Okajima T, Kuroda S, Nakamoto T, Taki M, Yamamoto Y, Hayashi H, Tanizawa K. Quantum mechanical hydrogen tunneling in bacterial copper amine oxidase reaction. Biochem Biophys Res Commun. 2006 Apr 7;342(2):414-23. Epub 2006 Feb 8. PMID:16487484 doi:10.1016/j.bbrc.2006.01.150
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Raymond Lyle, Alexander Berchansky, OCA, Jaime Prilusky