Sandbox Reserved 321
From Proteopedia
| Line 8: | Line 8: | ||
by Kelly Hrywkiw | by Kelly Hrywkiw | ||
{{STRUCTURE_2h9i | PDB=2h9i | SCENE= }} | {{STRUCTURE_2h9i | PDB=2h9i | SCENE= }} | ||
| + | [[Image:Secondary Structure of inhA.png|thumb|left|Width200Height300|alt=Secondary Structure Succession of inhA. Secondary structure residues are ordered from blue to red.|Secondary structure succession inhA.]] | ||
__TOC__ | __TOC__ | ||
| - | |||
| - | [[Image:Secondary Structure of inhA.png|thumb|left|Width200Height300|alt=Secondary Structure Succession of inhA. Secondary structure residues are ordered from blue to red.|Secondary structure succession inhA.]] | ||
=Introduction= | =Introduction= | ||
| - | The enzyme inhA is coded from the inhA gene that is simillar in sequence to the ''[http://en.wikipedia.org/wiki/Salmonella_typhimurium Salmonella typhimurium]''gene which plays a role in fatty acid biosynthesis <ref name ="making drugs for inhA">Sacchettini, James (New Rochelle, NY) 1999 INHA crystals and three dimensional structure United States Albert Einstein College of Medicine of Yeshiva University (Bronx, NY) 5882878 http://www.freepatentsonline.com/5882878.html</ref>. Inha is an NADH dependent trans enoyl-acyl ACP carrier protein that plays a role in the sysnthesis of [http://en.wikipedia.org/wiki/Mycolic_acid Mycolic Acid], and is part of a short-chain dehydrogenase/reductase family <ref name ="mech of thioamide drug action">PMID:17227913</ref><ref name ="phosphorylation of inhA">PMID:21143326</ref>. Mycolic acids are long chain fatty acids that are essential in cell wall formation of the human pathogen ''[http://en.wikipedia.org/wiki/Mycobacterium_tuberculosis Mycobacterium tuberculosis]''as well as other mycobateria such as ''[http://en.wikipedia.org/wiki/Mycobacterium_leprae Mycobacterium leprae]''<ref name ="TB">PMID2568869:</ref>. Inha has been propsed as the target of the | + | The enzyme inhA is coded from the inhA gene that is simillar in sequence to the ''[http://en.wikipedia.org/wiki/Salmonella_typhimurium Salmonella typhimurium]''gene which plays a role in fatty acid biosynthesis <ref name ="making drugs for inhA">Sacchettini, James (New Rochelle, NY) 1999 INHA crystals and three dimensional structure United States Albert Einstein College of Medicine of Yeshiva University (Bronx, NY) 5882878 http://www.freepatentsonline.com/5882878.html</ref>. Inha is an NADH dependent trans enoyl-acyl ACP carrier protein that plays a role in the sysnthesis of [http://en.wikipedia.org/wiki/Mycolic_acid Mycolic Acid], and is part of a short-chain dehydrogenase/reductase family <ref name ="mech of thioamide drug action">PMID:17227913</ref><ref name ="phosphorylation of inhA">PMID:21143326</ref>. Mycolic acids are long chain fatty acids that are essential in cell wall formation of the human pathogen ''[http://en.wikipedia.org/wiki/Mycobacterium_tuberculosis Mycobacterium tuberculosis]''as well as other mycobateria such as ''[http://en.wikipedia.org/wiki/Mycobacterium_leprae Mycobacterium leprae]''<ref name ="TB">PMID2568869:</ref>. Inha has been propsed as the target of the [http://en.wikipedia.org/wiki/Thioamidedrugs thioamide] drugs, ethionamide (ETH) and isoniazid (INH), which have been used in treatment of mycobacterial infections <ref name ="phosphorylation of inhA">PMID:21143326</ref>. |
=Structure of inhA= | =Structure of inhA= | ||
| - | <Structure load='2h9i' size=' | + | The inhA enzyme of ''M. tuberculosis'' is a homotetramer composed of a repeating subunit comprised of a single domain with a [http://en.wikipedia.org/wiki/Rossmann_fold Rossmann Fold] in the core that provides a NADH binding site<ref name ="crystallographic studies"/>. The single domain can be broken down into two substructures that are connected by short peptide loop<ref name ="making drugs for inhA"/><ref name ="crystallographic studies">PMID:17588773</ref>. |
| + | |||
| + | <Structure load='2h9i' size='250' frame='true' align='left' caption='Momomeric subunit of inhA' scene='Sandbox_Reserved_321/Structural_progresion/1' /> | ||
| - | The inhA enzyme of ''[http://en.wikipedia.org/wiki/Mycobacterium_tuberculosis Mycobacterium tuberculosis]'' is a homotetramer composed of a repeating subunit comprised of a single domain with a [http://en.wikipedia.org/wiki/Rossmann_fold Rossmann Fold] in the core that provides a NADH binding site<ref name ="crystallographic studies"/>. The single domain can be broken down into two substructures that are connected by short peptide loop<ref name ="making drugs for inhA"/><ref name ="crystallographic studies">PMID:17588773</ref>. | ||
==Substructure 1 of inhA== | ==Substructure 1 of inhA== | ||
| Line 28: | Line 28: | ||
The first substructure can be further broken down into two sections, the <scene name='Sandbox_Reserved_321/Substructure1section1/7'>first section</scene> consisting of two β strands <scene name='Sandbox_Reserved_321/B-1_and_b-2/4'>(B-1 and B-2)</scene>and two short α helicies <scene name='Sandbox_Reserved_321/A-1_and_a-2/2'>(A-1 and A-2)</scene><ref name ="making drugs for inhA"/>. | The first substructure can be further broken down into two sections, the <scene name='Sandbox_Reserved_321/Substructure1section1/7'>first section</scene> consisting of two β strands <scene name='Sandbox_Reserved_321/B-1_and_b-2/4'>(B-1 and B-2)</scene>and two short α helicies <scene name='Sandbox_Reserved_321/A-1_and_a-2/2'>(A-1 and A-2)</scene><ref name ="making drugs for inhA"/>. | ||
The first section is connected to the <scene name='Sandbox_Reserved_321/Section2substructure1/1'>second section</scene> by a β strand <scene name='Sandbox_Reserved_321/B-3/1'>(B-3)</scene> that crosses over the two domains, and leads into the second section initiating at the third α helix <scene name='Sandbox_Reserved_321/A-3/1'>(A-3)</scene><ref name ="making drugs for inhA"/>(A-3) is connected by a long loop to a 14 residue β strand <scene name='Sandbox_Reserved_321/B-4/2'>(B-4)</scene>that then leads into the fourth α helix <scene name='Sandbox_Reserved_321/A-4/2'>(A-4)</scene><ref name ="making drugs for inhA"/>. A-4 then leads into a fifth strand β <scene name='Sandbox_Reserved_321/B-5/1'>(B-5)</scene>, followed by a 25 residue α helix <scene name='Sandbox_Reserved_321/A-5/2'>(A-5)</scene>, and into the final strand β <scene name='Sandbox_Reserved_321/B-6/1'>(B-6)</scene><ref name ="making drugs for inhA"/>. | The first section is connected to the <scene name='Sandbox_Reserved_321/Section2substructure1/1'>second section</scene> by a β strand <scene name='Sandbox_Reserved_321/B-3/1'>(B-3)</scene> that crosses over the two domains, and leads into the second section initiating at the third α helix <scene name='Sandbox_Reserved_321/A-3/1'>(A-3)</scene><ref name ="making drugs for inhA"/>(A-3) is connected by a long loop to a 14 residue β strand <scene name='Sandbox_Reserved_321/B-4/2'>(B-4)</scene>that then leads into the fourth α helix <scene name='Sandbox_Reserved_321/A-4/2'>(A-4)</scene><ref name ="making drugs for inhA"/>. A-4 then leads into a fifth strand β <scene name='Sandbox_Reserved_321/B-5/1'>(B-5)</scene>, followed by a 25 residue α helix <scene name='Sandbox_Reserved_321/A-5/2'>(A-5)</scene>, and into the final strand β <scene name='Sandbox_Reserved_321/B-6/1'>(B-6)</scene><ref name ="making drugs for inhA"/>. | ||
| + | |||
==Substructure 2 of inhA== | ==Substructure 2 of inhA== | ||
| Line 33: | Line 34: | ||
<scene name='Sandbox_Reserved_321/Substructure_2/1'>Substructure 2</scene> contains the c-terminal region of the molecule and consists of a small β strand <scene name='Sandbox_Reserved_321/B-7/1'>(B-7)</scene>, and two α helicies <scene name='Sandbox_Reserved_321/A-6_and_a-7/1'>(A-6 and A-7)</scene> which are conected by a short five residue loop<ref name ="making drugs for inhA"/>. The C-terminal domain consits of two other α helicies <scene name='Sandbox_Reserved_321/A-8_and_a-9/1'>(A-8 and A-9)</scene><ref name ="making drugs for inhA"/>. | <scene name='Sandbox_Reserved_321/Substructure_2/1'>Substructure 2</scene> contains the c-terminal region of the molecule and consists of a small β strand <scene name='Sandbox_Reserved_321/B-7/1'>(B-7)</scene>, and two α helicies <scene name='Sandbox_Reserved_321/A-6_and_a-7/1'>(A-6 and A-7)</scene> which are conected by a short five residue loop<ref name ="making drugs for inhA"/>. The C-terminal domain consits of two other α helicies <scene name='Sandbox_Reserved_321/A-8_and_a-9/1'>(A-8 and A-9)</scene><ref name ="making drugs for inhA"/>. | ||
| - | = | + | |
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | =Function= | ||
| + | |||
| + | InhA plays a key role in the synthesis of fatty acids, particularly in ''M. tuberculosis'' which has type one fatty acid synthesis (FASI) and type two fatty acid synthesis (FASII) which together funtion in the synthesis of mycolic acids.<ref name ="Function of M Tb">PMID:18552191</ref>. The final step in FASII is compleated by InhA which reduces 2-trans-enoyl-ACP's with chain lengths over twelve carbons <ref name ="Function of M Tb"/> | ||
=Role in the Mycolic Acid Pathway= | =Role in the Mycolic Acid Pathway= | ||
| - | [[Image:Pathway.png|thumb|right|upright=2|alt=Proposed mechanism.|Formulated mechanism of Mycolic acid synthesis as proposed by Wilson et al.<ref name ="Drug Induced Alterations">10536008 </ref>.]] | + | [[Image:Pathway.png|thumb|right|upright=2|alt=Proposed mechanism.|Formulated mechanism of Mycolic acid synthesis as proposed by Wilson et al.<ref name ="Drug Induced Alterations">PMID:10536008</ref>.]] |
| Line 44: | Line 53: | ||
=References= | =References= | ||
| - | |||
| - | <references/> | ||
Revision as of 01:02, 1 April 2011
| This Sandbox is Reserved from January 10, 2010, through April 10, 2011 for use in BCMB 307-Proteins course taught by Andrea Gorrell at the University of Northern British Columbia, Prince George, BC, Canada. |
To get started:
More help: Help:Editing |
InhA
by Kelly Hrywkiw
| |||||||||
| 2h9i, resolution 2.20Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | |||||||||
| Gene: | inhA (Mycobacterium tuberculosis) | ||||||||
| Activity: | [acyl-carrier-protein_reductase_(NADH) Enoyl-[acyl-carrier-protein] reductase (NADH)], with EC number 1.3.1.9 | ||||||||
| Related: | 1zid | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Contents |
Introduction
The enzyme inhA is coded from the inhA gene that is simillar in sequence to the Salmonella typhimuriumgene which plays a role in fatty acid biosynthesis [1]. Inha is an NADH dependent trans enoyl-acyl ACP carrier protein that plays a role in the sysnthesis of Mycolic Acid, and is part of a short-chain dehydrogenase/reductase family [2][3]. Mycolic acids are long chain fatty acids that are essential in cell wall formation of the human pathogen Mycobacterium tuberculosisas well as other mycobateria such as Mycobacterium leprae[4]. Inha has been propsed as the target of the thioamide drugs, ethionamide (ETH) and isoniazid (INH), which have been used in treatment of mycobacterial infections [3].
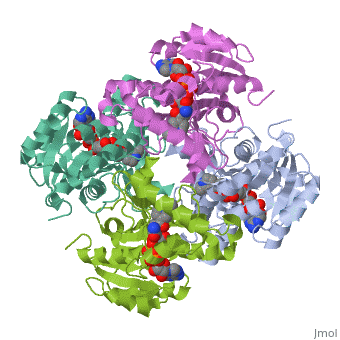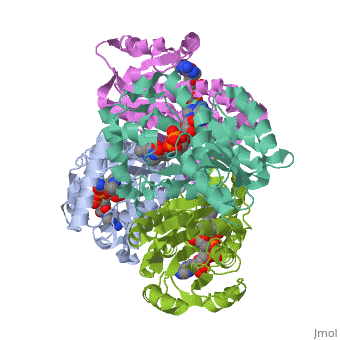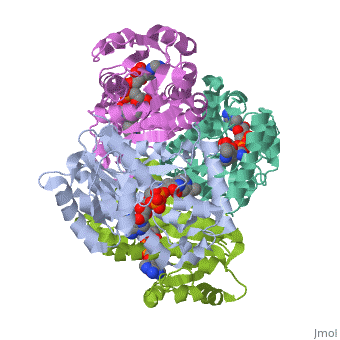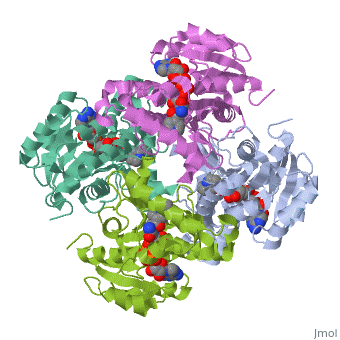
Structure of inhA
The inhA enzyme of M. tuberculosis is a homotetramer composed of a repeating subunit comprised of a single domain with a Rossmann Fold in the core that provides a NADH binding site[5]. The single domain can be broken down into two substructures that are connected by short peptide loop[1][5].
|
Substructure 1 of inhA
consists of 6 parallel β strands and 4 α helices interwoven together to form a core α/β structure that contains the n-terminal domain[1]. The first substructure can be further broken down into two sections, the consisting of two β strands and two short α helicies [1]. The first section is connected to the by a β strand that crosses over the two domains, and leads into the second section initiating at the third α helix [1](A-3) is connected by a long loop to a 14 residue β strand that then leads into the fourth α helix [1]. A-4 then leads into a fifth strand β , followed by a 25 residue α helix , and into the final strand β [1].
Substructure 2 of inhA
contains the c-terminal region of the molecule and consists of a small β strand , and two α helicies which are conected by a short five residue loop[1]. The C-terminal domain consits of two other α helicies [1].
Function
InhA plays a key role in the synthesis of fatty acids, particularly in M. tuberculosis which has type one fatty acid synthesis (FASI) and type two fatty acid synthesis (FASII) which together funtion in the synthesis of mycolic acids.[6]. The final step in FASII is compleated by InhA which reduces 2-trans-enoyl-ACP's with chain lengths over twelve carbons [6]
Role in the Mycolic Acid Pathway