Ribonuclease A Catalysis
From Proteopedia
| Line 12: | Line 12: | ||
===Structure=== | ===Structure=== | ||
| - | The catalysis of RNA strands occurs in the active site which is the location of the chemical change in bound substrates. Subsites both up and down stream of the residues found around the active site are important for the binding of single stranded RNA. Large | + | The catalysis of RNA strands occurs in the active site which is the location of the chemical change in bound substrates. Subsites both up and down stream of the residues found around the active site are important for the binding of single stranded RNA. Large quantities of positively charged residues, such as Lys 7 and 66 and Arg 10, attract the negative charge on the phosphate back bone of the RNA strand. |
=='''Acid Base Catalysis by RNase A'''== | =='''Acid Base Catalysis by RNase A'''== | ||
| - | In the acid base catalysis of RNA in mammalians, RNase A catalyzes the cleavage of the P-O 5’ bond, and is comprised of two separate processes, the formation of the | + | In the acid base catalysis of RNA in mammalians, RNase A catalyzes the cleavage of the P-O 5’ bond, and is comprised of two separate processes, the formation of the pentavalent phosphate transition state and subsequent degradation 2’3’ cyclic intermediate into its individual nucleotides. An important part of the reaction is Histidine’s ability to both accept and donate electrons. This acts as a proton source, allowing Histidine to be utilized as a base or acid, making the reaction pH dependent. |
Revision as of 02:23, 1 April 2011
| This Sandbox is Reserved from Feb 02, 2011, through Jul 31, 2011 for use by the Biochemistry II class at the Butler University at Indianapolis, IN USA taught by R. Jeremy Johnson. This reservation includes Sandbox Reserved 191 through Sandbox Reserved 200. |
To get started:
More help: Help:Editing |
Contents |
Ribonuclease A Catalysis
Introduction
| |||||||||
| 7rsa, resolution 1.26Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , | ||||||||
| Activity: | Pancreatic ribonuclease, with EC number 3.1.27.5 | ||||||||
| |||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Acid Base Catalysis
Acid Base Catalysis is the acceleration of a chemical reaction by the addition of an acid or a base and is mainly used in organic chemical reactions. The acid or base is not consumed in the reaction itself. An acid transfers protons to a reactant and a base accepts protons from the reactant. An acid is often thought of as a proton and the base as a hydroxyl. When the acids or bases donate or accept protons, they stabilize the developing charges in the transition state. This usually creates a better leaving group, making the reaction more energetically favorable. Additionally, this has an effect on the activity of the nucleophile and electrophile groups. Histidine is a very common residue involved in acid-base cataylsis due to the fact that is has a pKa close to neutral; therefore, it can both accept and donate protons.
The acid base mechanism can extensively alter the pKa depending on the environment of the residue. PKa will increase for an acidic residue if the environment is hydrophobic or if the adjacent residues are of similar charges. In the same environmental conditions, a basic residue will decrease the pKa. PKa will decrease for an acidic residue and increase for a basic residue if there is a salt bridge.
Structure
The catalysis of RNA strands occurs in the active site which is the location of the chemical change in bound substrates. Subsites both up and down stream of the residues found around the active site are important for the binding of single stranded RNA. Large quantities of positively charged residues, such as Lys 7 and 66 and Arg 10, attract the negative charge on the phosphate back bone of the RNA strand.
Acid Base Catalysis by RNase A
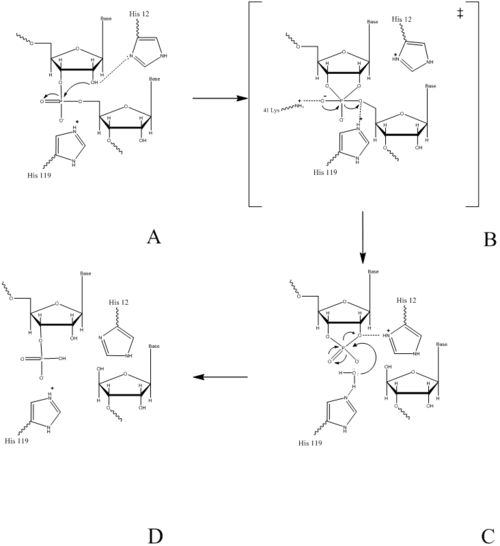
In the acid base catalysis of RNA in mammalians, RNase A catalyzes the cleavage of the P-O 5’ bond, and is comprised of two separate processes, the formation of the pentavalent phosphate transition state and subsequent degradation 2’3’ cyclic intermediate into its individual nucleotides. An important part of the reaction is Histidine’s ability to both accept and donate electrons. This acts as a proton source, allowing Histidine to be utilized as a base or acid, making the reaction pH dependent.
The catalysis of RNase A begins when His-12 undergoes basic catalysis. H-12 will act as a base and abstract a proton from the RNA’s 2’ OH group; thus, assisting the attack of the 2’ oxygen on the phosphorus atom. This reaction occurs via a transition state, having a pentavalent phosphorous atom. These transition states are both stabilized by the side positive character of the side chain of Lys41 and the main chain of Phe120. This leads to the formation a stabilized 2’3’-cyclic intermediate. His-119 will support this reaction by protonating the leaving group, the 6’ OH on the ribose of the 3’ RNA, thus acting as a general acid.
The 2’,3’- cyclic nucleotide is hydrolyzed in a separate process. His-12 will donate the excess proton from the initial step to the leaving group, the 3’ oxygen of the cyclic intermediate. Simultaneously, His-119 draws the hydrogen off of a water molecule. His 119 is thus reprotonated during this process, making water a better nucleophile. The water molecule attacks the phosphate causing the cleavage of the 2-3’ cyclic intermediate. The truncated nucleotide is then released with a 3’ phosphate group. Upon degredation of the phosphodiester linkage between the two nucleotides the products are then released into the surrounding solvent.
Related Sites
Works Cited
1.Raines, R. Ribonuclease A. Chemistry Review: (1998) Vol. 98 pp. 1045-1068
2.Wlodrawer, A., Svensson, L., Sjohin, L., Gilliland, G. Structure of Phosphate-Free Ribonuclease A Refined at 1.26A. Biochemistry:(1988) Vol. 27 pp. 2705-2717
Proteopedia Page Contributors and Editors (what is this?)
Nathan Clarke, David Canner, Alexander Berchansky, R. Jeremy Johnson, OCA