Sandbox Reserved 338
From Proteopedia
| Line 13: | Line 13: | ||
===Structure and Function=== | ===Structure and Function=== | ||
| - | <Structure load='2vnc' size='300' frame='true' align='right | + | <Structure load='2vnc' size='300' frame='true' align='right' scene='Sandbox_Reserved_338/2vnc/1'> |
TreX is an oligomer, as it exists in a dimeric state and a tetrameric state, both of which exhibit different enzymatic activities. All subunits are identical, where the monomer contains a total of 612 amino acids <ref name="Woo" />. The polypeptide folds into two secondary structures, a β-sandwhich in the N terminal region, comprised of six β-strands and a (β/α)8 – barrel motif in the central domain, comprised of eight parallel α-strands which encircle eight parallel β-strands. The sequence composition of the TreX monomer exhibits a high degree of homology to the isoamylase debranching enzyme of Pseudomona, however the TreX monomer mainly deviates from this similarity in its substrate binding groove and the absence of a calcium ion ligand <ref name="Woo" />. | TreX is an oligomer, as it exists in a dimeric state and a tetrameric state, both of which exhibit different enzymatic activities. All subunits are identical, where the monomer contains a total of 612 amino acids <ref name="Woo" />. The polypeptide folds into two secondary structures, a β-sandwhich in the N terminal region, comprised of six β-strands and a (β/α)8 – barrel motif in the central domain, comprised of eight parallel α-strands which encircle eight parallel β-strands. The sequence composition of the TreX monomer exhibits a high degree of homology to the isoamylase debranching enzyme of Pseudomona, however the TreX monomer mainly deviates from this similarity in its substrate binding groove and the absence of a calcium ion ligand <ref name="Woo" />. | ||
Revision as of 00:49, 2 April 2011
| This Sandbox is Reserved from January 10, 2010, through April 10, 2011 for use in BCMB 307-Proteins course taught by Andrea Gorrell at the University of Northern British Columbia, Prince George, BC, Canada. |
To get started:
More help: Help:Editing |
| |||||||||
| 2vnc, resolution 3.00Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Related: | 2vuy, 2vr5, 2vnb | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Introduction
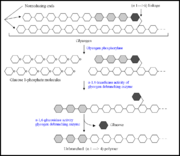
A glycogen-debranching enzyme (GDE) is one of the enzymes associated with the breakdown of glycogen [1]. The majority of debranching enzymes belong to the GH13 (glycoside hydrolase 13) family [2], and they may further be separated into two main groups based on function [1]. For example, in mammals and yeast, the GDE possesses two functions; both α-1,6-glycosidase and α-1,4-transferase activity, and thus catalyzes two successive reactions in the transfer of glycogen branches [1]. In bacteria and plants, however, the debranching of glycogen is carried out by two different enzymes, where one possess α-1,6-glycosidase activity and the other possesses α-1,4-transferase activity, but not both [3]. For example, isoamylases and pullulanases carry out the α-1,6-glycosidic bond hydrolyzing activity [4], while glucosyltransferases carry out the α-1,4-transferase activity [2].
TreX
TreX is an archaeal GDE from the species, Sulfolobus solfataricus [1]. Interestingly, TreX exhibits 74% sequence similarity to the isoamylase from Sulfolobus acidocaldarium, yet TreX itself reveals both α-1,6-glycosidase and α-1,4-transferase activity. It functions to debranch the side chains of glycogen into maltodextrin, and subsequently TreY and TreZ convert the maltodextrin into trehalose [1] [5]. Although TreX exhibits bifunctional activity, its catalytic region differs greatly from other glycogen debranching enzymes. For example, human and yeast GDEs have distinct catalytic sites for the α-1,6-glycosidase activity and α-1,4-transferase activity. These sites are even located at different regions of the polypeptide. In TreX, however, both enzymatic rections take place within the same catalytic region [1].
Structure and Function
| |||||||||||