Sandbox Reserved 194
From Proteopedia
| Line 3: | Line 3: | ||
<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | <!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | ||
== '''Ribonuclease A Substrate Binding''' == | == '''Ribonuclease A Substrate Binding''' == | ||
| - | |||
== Introduction == | == Introduction == | ||
| + | <Structure load='1RTA' size='450' frame='true' align='right' caption='Thymidylic acid tetramer and ApTpApApG complexed with ribonuclease A showing pi stacking and hydrogen bonding ' scene='Sandbox_Reserved_194/1rta_structure/2'/> | ||
| - | + | RNase A is an endonuclease that cleaves and breaks down RNA using acid base catalysis (link to sandbox 193). RNase A has been a model protein for studies on the stability, folding and chemistry of proteins. ‘<ref>PMID:11848924</ref>’ It is also essential in protein regulation within the body due to its function of RNA degradation. | |
| - | + | ||
| - | + | ||
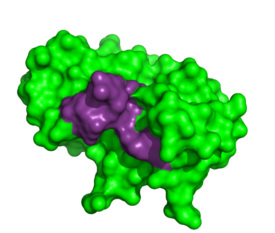
[[Image:1RTAnew.png|thumb|left|275px|Thymidylic acid tetramer complexed with ribonuclease A]] | [[Image:1RTAnew.png|thumb|left|275px|Thymidylic acid tetramer complexed with ribonuclease A]] | ||
| Line 16: | Line 14: | ||
== Substrate Binding == | == Substrate Binding == | ||
| - | To determine the structural characteristics of RNA substrate binding to RNase A, X-ray crystallography was used to image inhibitory DNA tetramers bound to | + | To determine the structural characteristics of RNA substrate binding to RNase A, X-ray crystallography was used to image inhibitory DNA tetramers bound to RNase A. DNA lacks the 2’OH essential to RNA cleavage, making the complex more conducive to crystallography. The complex between RNase A and thymidylic acid tetramer (d(pT)4) provides information about specificity of the binding pocket subunits, B0, B1, B2 and B3. Many interactions observed in this complex occur between amino acid residues and the nucleic acid backbone. Examples of these interactions include hydrogen bonding between <scene name='Sandbox_Reserved_194/1rta_structure_arg/4'> phosphate of thymine 1 and Arg39</scene> as well as hydrogen bonding between the O5’ oxygen of the ribose of thymine 3 and Lys41. ‘<ref>PMID:1429575</ref>’ |
| - | + | ||
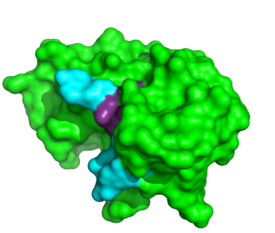
[[Image:1RCNnew.png|thumb|left|275px|ApTpApApG complexed with ribonuclease A]] | [[Image:1RCNnew.png|thumb|left|275px|ApTpApApG complexed with ribonuclease A]] | ||
| + | Further binding pocket characterization was performed using the oligonucleotides <scene name='Sandbox_Reserved_194/1rcn_structure/3'>d(ApTpApApG)</scene>. In this complex, the B1 site is thought to exclusively bind to pyrimidine bases due to steric interactions and hydrogen bonding to Thr45. When Thr45 was mutated to glycine, purines readily bound to the B1 site. ‘<ref>PMID: 8193116</ref>’ This interaction appears to be the driving force behind its inability to bind purines. While binding of other nucleobases to the B2 and B3 sites is possible, the imaging of this complex elucidated the preferences for adenosine bases at these two positions. In addition to its catalytic activity His119 has also been shown to be important in substrate specificity. This is due to the <scene name='Sandbox_Reserved_194/1rcn_his/9'>pi stacking between His119 and A3 </scene>. When this site was mutated, the affinity for a poly(A) substrate was decreased by 104-fold. ‘<ref>PMID: 21391696</ref>It also establishes hydrogen bonding between <scene name='Sandbox_Reserved_194/1rcn_hydrogen_bonding/9'>Asn71-A3, Gln69-A3 and Gln69-A4</scene>. ‘<ref>PMID:8063789</ref>’ | ||
| - | + | == Conclusion == | |
| - | + | ||
| - | + | ||
| - | + | From the studies on RNase A complexes with deoxy nucleic acid tetramers, it has been established that this enzyme recognizes the substrate on both its phosphate backbone and on individual nucleobases. RNase A has a nonspecific B0 site, a B1 site specific to pyrimidines and a B3 and B4 site with a preference for adenosine bases. Similar to other enzymes, RNase A uses the hydrogen bonding distance between amino acids and the substrate to bind specifically to certain nucleobases. Studying the substrate recognition and specificity of enzymes such as RNase A is an important step in understanding the regulation of RNA within biological systems. | |
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 01:54, 15 April 2011
| This Sandbox is Reserved from Feb 02, 2011, through Jul 31, 2011 for use by the Biochemistry II class at the Butler University at Indianapolis, IN USA taught by R. Jeremy Johnson. This reservation includes Sandbox Reserved 191 through Sandbox Reserved 200. |
To get started:
More help: Help:Editing |
Contents |
Ribonuclease A Substrate Binding
Introduction
|
RNase A is an endonuclease that cleaves and breaks down RNA using acid base catalysis (link to sandbox 193). RNase A has been a model protein for studies on the stability, folding and chemistry of proteins. ‘[1]’ It is also essential in protein regulation within the body due to its function of RNA degradation.
Substrate Binding
To determine the structural characteristics of RNA substrate binding to RNase A, X-ray crystallography was used to image inhibitory DNA tetramers bound to RNase A. DNA lacks the 2’OH essential to RNA cleavage, making the complex more conducive to crystallography. The complex between RNase A and thymidylic acid tetramer (d(pT)4) provides information about specificity of the binding pocket subunits, B0, B1, B2 and B3. Many interactions observed in this complex occur between amino acid residues and the nucleic acid backbone. Examples of these interactions include hydrogen bonding between as well as hydrogen bonding between the O5’ oxygen of the ribose of thymine 3 and Lys41. ‘[2]’
Further binding pocket characterization was performed using the oligonucleotides . In this complex, the B1 site is thought to exclusively bind to pyrimidine bases due to steric interactions and hydrogen bonding to Thr45. When Thr45 was mutated to glycine, purines readily bound to the B1 site. ‘[3]’ This interaction appears to be the driving force behind its inability to bind purines. While binding of other nucleobases to the B2 and B3 sites is possible, the imaging of this complex elucidated the preferences for adenosine bases at these two positions. In addition to its catalytic activity His119 has also been shown to be important in substrate specificity. This is due to the . When this site was mutated, the affinity for a poly(A) substrate was decreased by 104-fold. ‘[4]It also establishes hydrogen bonding between . ‘[5]’
Conclusion
From the studies on RNase A complexes with deoxy nucleic acid tetramers, it has been established that this enzyme recognizes the substrate on both its phosphate backbone and on individual nucleobases. RNase A has a nonspecific B0 site, a B1 site specific to pyrimidines and a B3 and B4 site with a preference for adenosine bases. Similar to other enzymes, RNase A uses the hydrogen bonding distance between amino acids and the substrate to bind specifically to certain nucleobases. Studying the substrate recognition and specificity of enzymes such as RNase A is an important step in understanding the regulation of RNA within biological systems.
References
- ↑ Raines RT. Ribonuclease A. Chem Rev. 1998 May 7;98(3):1045-1066. PMID:11848924
- ↑ Birdsall DL, McPherson A. Crystal structure disposition of thymidylic acid tetramer in complex with ribonuclease A. J Biol Chem. 1992 Nov 5;267(31):22230-6. PMID:1429575
- ↑ delCardayre SB, Raines RT. Structural determinants of enzymatic processivity. Biochemistry. 1994 May 24;33(20):6031-7. PMID:8193116
- ↑ Thompson JE, Raines RT. Value of general Acid-base catalysis to ribonuclease a. J Am Chem Soc. 1994 Jun;116(12):5467-8. PMID:21391696 doi:10.1021/ja00091a060
- ↑ Fontecilla-Camps JC, de Llorens R, le Du MH, Cuchillo CM. Crystal structure of ribonuclease A.d(ApTpApApG) complex. Direct evidence for extended substrate recognition. J Biol Chem. 1994 Aug 26;269(34):21526-31. PMID:8063789