Sandbox Mati
From Proteopedia
| Line 8: | Line 8: | ||
==Formation of factor XIa via factor XI zymogen activation== | ==Formation of factor XIa via factor XI zymogen activation== | ||
| - | Factor XI is partially proteolyzed ''in vitro'' by thrombin and factor XIIa generating the active serine-protease, factor XIa. Similar to other chymotrypsin-like proteases, its topology consist of two β-barrels linked through a central loop. Next to the C-terminal | + | Factor XI is partially proteolyzed ''in vitro'' by thrombin and factor XIIa generating the active serine-protease, factor XIa. Similar to other chymotrypsin-like proteases, its topology consist of two β-barrels linked through a central loop. Next to the C-terminal Cys-356 of the factor XI heavy chain, the polypeptide forms a 3-10 helix conformation and again turn sharply 90 degrees at Cys-362 forming a disulfide bond with Cys-482 within the active site region. Thrombin-catalyzed proteolysis of factor XI involves crucial interations with Glu-66, Lys-83 and Gln-84 of the A1 domain(this ensures maximum proximity to the activation loop of factor XI)of the factor XI molecule through its exosites I and II regions <ref>PMID:16699514</ref>. Thus binding of thrombin to one subunit of the zymogen dimer promotes cleavage of the bond between Arg369-Ile370 contained in the activation loop of factor XI. The activation loop (residues 366-370) consequently undergoes the greatest conformational change as Ile-370 is displaced ~20Å from its position in factor XI and inserts into the activation pocket of factor XIa producing the oxyanion hole in the active site of the protease <ref>PMID:14523451</ref>. |
| - | ==Active Site | + | ==Active Site== |
| - | + | Similar to other serine proteases, the [http://en.wikipedia.org/wiki/Catalytic_triad catalytic triad] residues Ser-557, Asp-462 and His-413 constitute the <scene name='Sandbox/Active_site/1'>active site</scene> of factor XIa. A [http://en.wikipedia.org/wiki/Low-barrier_hydrogen_bond low barrier hydrogen bond (LBHB)] formed between the carboxyl group of Asp-462 and imidazole nitrogen of His 413 causes the deprotonation Ser-557 (enhacing its nucleophilicity). Thus catalysis involves a nucleophilic attack by Ser-557 on the carbonyl carbon of the target amino acid at the C-terminal of the substrate producing an intermediate which is stablized by the oxyanion hole. Rearrangement of the resulting tetrahedral intermediate and a second nucleophilic attack by water yields a cleaved peptide with a free carboxyl end <ref>PMID:14523451</ref>. | |
| - | <Structure load='3bg8' size='400' frame='true' align='right' caption='Factor XIa light chain | + | <Structure load='3bg8' size='400' frame='true' align='right' caption='Factor XIa light chain' /> |
Revision as of 23:11, 17 April 2011
Contents |
Coagulation Factor XIa
Introduction
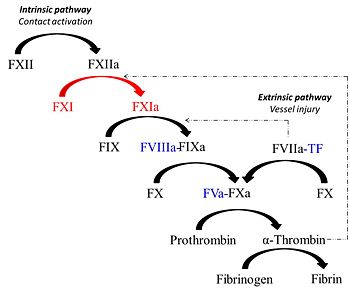
Factor XIa is unique protease derived from the activation of the coagulation zymogen, factor XI. Factor XIa partcipates in the procoagulant response via contact activation pathway. Synthesized by the liver similar to most vitamin K-dependent coagulation proteins, the zymogen, factor XI circulates in plasma as a 160 kDa disulfide-linked homodimer in complex with high molecular weight kininogen (HK)[1]. Studies show that factor XI is a substrate for various plasma proteins such as factor XIIa, thrombin, meizothrombin and factor XIa (via autoactivation). Proteolysis of the Arg369-Ile370 bond generates the active enzyme factor XIa which in turn cleaves its substrate factor factor IX to produce the serine protease factor IXa.
Protein Structure
Each dimeric subunit of factor XIa exhibit similar amino acid composition of about 607 residues constituting five main domains. The N-terminus of each subunit contains 4 apple domains (A1, A2, A3 and A4) which are characterized by approximately 90 or 91 amino acid repeats. The C-terminus contain the trypsin-like catalytic domain. Together with Prekallikrein (PK) a monomeric homolog of factor XIa, they belong to the PAN (plasminogen, apple, nematode) module family which all have a conserved N-terminal apple domain )[2]. The topology of the apple domain reveals 7 antiparallel β-sheets and an α-helix which fold into a compact structure as oppose to an extended structure found in the vitamin K-dependent serine proteases. This core PAN topology is also found in leech antiplatelet protein and hepatocyte growth factor)[3]. A single disulfide linkage connects the C- and N-terminals of the dimer whereas two disulfide bond join the heix to the 4β- and 5β-sheets. The apple domains of Factor XIa are tightly linked to each other forming a disk-like structure closed to the base of the C-terminal catalytic domain. This observation is consistent with the high surface area measurements for the side interfaces between apple domains A1 and A2 (441ÅxÅ) and between A3 and A4 (444ÅxÅ) in contrast to smaller end interfaces between A1 and A4(380ÅxÅ) and between A2 and A3(284ÅxÅ).
Formation of factor XIa via factor XI zymogen activation
Factor XI is partially proteolyzed in vitro by thrombin and factor XIIa generating the active serine-protease, factor XIa. Similar to other chymotrypsin-like proteases, its topology consist of two β-barrels linked through a central loop. Next to the C-terminal Cys-356 of the factor XI heavy chain, the polypeptide forms a 3-10 helix conformation and again turn sharply 90 degrees at Cys-362 forming a disulfide bond with Cys-482 within the active site region. Thrombin-catalyzed proteolysis of factor XI involves crucial interations with Glu-66, Lys-83 and Gln-84 of the A1 domain(this ensures maximum proximity to the activation loop of factor XI)of the factor XI molecule through its exosites I and II regions [4]. Thus binding of thrombin to one subunit of the zymogen dimer promotes cleavage of the bond between Arg369-Ile370 contained in the activation loop of factor XI. The activation loop (residues 366-370) consequently undergoes the greatest conformational change as Ile-370 is displaced ~20Å from its position in factor XI and inserts into the activation pocket of factor XIa producing the oxyanion hole in the active site of the protease [5].
Active Site
Similar to other serine proteases, the catalytic triad residues Ser-557, Asp-462 and His-413 constitute the of factor XIa. A low barrier hydrogen bond (LBHB) formed between the carboxyl group of Asp-462 and imidazole nitrogen of His 413 causes the deprotonation Ser-557 (enhacing its nucleophilicity). Thus catalysis involves a nucleophilic attack by Ser-557 on the carbonyl carbon of the target amino acid at the C-terminal of the substrate producing an intermediate which is stablized by the oxyanion hole. Rearrangement of the resulting tetrahedral intermediate and a second nucleophilic attack by water yields a cleaved peptide with a free carboxyl end [6].
|
Recognition of Substrates and Cleavege Mechanism
References
- ↑ Thompson RE, Mandle R Jr, Kaplan AP. Association of factor XI and high molecular weight kininogen in human plasma. J Clin Invest. 1977 Dec;60(6):1376-80. PMID:915004 doi:http://dx.doi.org/10.1172/JCI108898
- ↑ Tordai H, Banyai L, Patthy L. The PAN module: the N-terminal domains of plasminogen and hepatocyte growth factor are homologous with the apple domains of the prekallikrein family and with a novel domain found in numerous nematode proteins. FEBS Lett. 1999 Nov 12;461(1-2):63-7. PMID:10561497
- ↑ Tordai H, Banyai L, Patthy L. The PAN module: the N-terminal domains of plasminogen and hepatocyte growth factor are homologous with the apple domains of the prekallikrein family and with a novel domain found in numerous nematode proteins. FEBS Lett. 1999 Nov 12;461(1-2):63-7. PMID:10561497
- ↑ Papagrigoriou E, McEwan PA, Walsh PN, Emsley J. Crystal structure of the factor XI zymogen reveals a pathway for transactivation. Nat Struct Mol Biol. 2006 Jun;13(6):557-8. Epub 2006 May 14. PMID:16699514 doi:10.1038/nsmb1095
- ↑ Friedrich R, Panizzi P, Fuentes-Prior P, Richter K, Verhamme I, Anderson PJ, Kawabata S, Huber R, Bode W, Bock PE. Staphylocoagulase is a prototype for the mechanism of cofactor-induced zymogen activation. Nature. 2003 Oct 2;425(6957):535-9. PMID:14523451 doi:10.1038/nature01962
- ↑ Friedrich R, Panizzi P, Fuentes-Prior P, Richter K, Verhamme I, Anderson PJ, Kawabata S, Huber R, Bode W, Bock PE. Staphylocoagulase is a prototype for the mechanism of cofactor-induced zymogen activation. Nature. 2003 Oct 2;425(6957):535-9. PMID:14523451 doi:10.1038/nature01962