Fumarase 2
From Proteopedia
| Line 8: | Line 8: | ||
===Stucture and Classification=== | ===Stucture and Classification=== | ||
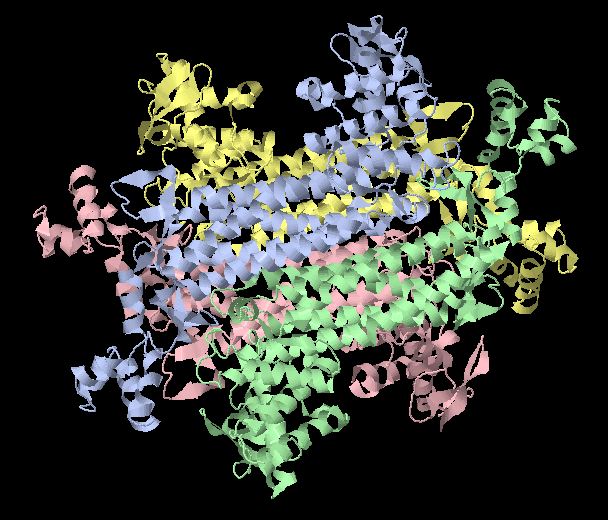
Fumarase is classified as an all alpha protein which belongs to the L-aspartase/fumarase family, and the enzyme specifically consists of four identical subunits which form a tetramer. From the four subunits, fumarase has three domains which comprise two binding sites: the active site and B site. Although the active site has a mostly solid structure and shifts very little when it binds, the B site shifts substantially more upon binding, and this shift helps regulate affinity for molecule binding at the active site (Weaver 2005). | Fumarase is classified as an all alpha protein which belongs to the L-aspartase/fumarase family, and the enzyme specifically consists of four identical subunits which form a tetramer. From the four subunits, fumarase has three domains which comprise two binding sites: the active site and B site. Although the active site has a mostly solid structure and shifts very little when it binds, the B site shifts substantially more upon binding, and this shift helps regulate affinity for molecule binding at the active site (Weaver 2005). | ||
| - | [[Image:Fumarase biological unit.JPG]] | + | <[[Image:Fumarase biological unit.JPG]] size='300' /> |
Revision as of 09:01, 28 April 2011
Contents |
Fumarase
Overview
Fumarase, also known as fumarate hydratase, functions as an enzyme in the metabolic pathway known as the Kreb’s cycle, or citric acid cycle. As the seventh step in the pathway, fumarase catalyzes the reversible reaction converting fumarate to S-malate. It metabolizes Fumarate in the cytosol, which becomes a byproduct of the urea cycle and amino acid catabolism. It catalyzes the addition of water to make S-Malate; therefore, the mechanism of fumarase in the reaction involves hydration of fumarate in order to form S-malate.
Stucture and Classification
Fumarase is classified as an all alpha protein which belongs to the L-aspartase/fumarase family, and the enzyme specifically consists of four identical subunits which form a tetramer. From the four subunits, fumarase has three domains which comprise two binding sites: the active site and B site. Although the active site has a mostly solid structure and shifts very little when it binds, the B site shifts substantially more upon binding, and this shift helps regulate affinity for molecule binding at the active site (Weaver 2005).
<
Mechanism of Reaction
Fumarase has the ability to catalyze the hydration of fumarate to malate or the dehydration of malate to fumarate. The mechanism of fumarase in the hydration and dehydration reaction pathways remains simple, only involving three steps. In the dehydration reaction, fumarase deprotonates a carbon atom on malate to form a carbanion (Beechmans 1998). This deprotonation results in an aci-carboxylate intermediate (Weaver 2005). After the intermediate forms, the acidic proton from the initial step removes the hydroxide group from the aci-carboxylate intermediate to form fumarase which then detaches from the active site of fumarase, completing the reaction (Rose, Weaver, 2004). In the active site, amino acid residues involved in binding the substrate are located on three subunits: Thr100, Ser139, Ser140, and Asn141 from the b-subunit, Thr187 and His188 on the d-subunit, and Lys324 and Asn326 of the c-subunit (Beechmans 198). The B site is located in a π-helix turn between the active site and solvent, and it includes residues Arg126, Lys127, Val128, His129, Pro130, Asn131, and Asp132 all on the b-subunit. Two hydrogen bonds initiate the binding of Asn131 and Asp132 residues with S-malate (Rose, Weaver, 2004).
| |||||||||||

