We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox89220
From Proteopedia
(Difference between revisions)
(→Structure) |
(→Structure) |
||
| Line 10: | Line 10: | ||
<Structure load='2amy' size='400' frame='true' align='right' scene='Sandbox89220/2amy/12'> caption='Phosphomannose mutase 2'/> | <Structure load='2amy' size='400' frame='true' align='right' scene='Sandbox89220/2amy/12'> caption='Phosphomannose mutase 2'/> | ||
| - | |||
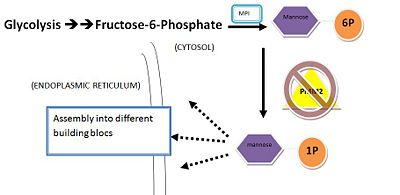
<scene name='Sandbox89220/2amy/12'>PMM2</scene> is a 246 amino acid long protein, with 2 identical subunits having close resemblance in their <scene name='Sandbox89220/2amy/17'>secondary structure</scene> working as independent domains, hence it’s also known as a homodimeric protein. Even if the structure of PMM2 is already solved,however, it's still not clear where its domains and its critical motifs are. Nonetheless, through the extraploration of PMM1, it’s known to be classified as the member of HAD (haloacid dehydrolase) superfamily due to the fact that it had 4 different types of motifs[http://www.sinauer.com/pdf/nsp-protein-1-16.pdf], each of which is highly conserved across the vertebrates’ lineages during evolutions. Similar to other members in HAD superfamily, the two domains are separately recognized as core and cap domain. The core domain (residues 1 – 90, and 198-262)of PMM2 displays the characteristic 4 motifs of HAD superfamily that contribute to the catalytic ability of the active sites in the protein. Motif 1 has <scene name='Sandbox89220/2amy/25'>Asp</scene> as nucleophile which serves as the mediator for the phosphoryl group transfer, the second Asp, on the other hand, acts as in the general acid-base reaction. While motif 1 provides the nucleophilic and general acid-base reactions, motif 2 in the protein which usually contains <scene name='Sandbox89220/2amy/26'>Thr</scene> or <scene name='Sandbox89220/2amy/28'>Ser</scene> helps in positioning or binding on the substrates’ phosphoryl group. In addition, motif 3 has the typical Lys or Arg residues while motif 4 has the acidic residues (Asp and Glu) that bind magnesium cofactor[http://en.wikipedia.org/wiki/Cofactor_%28biochemistry%29]<ref name="Evolutionary history">Rita Quental et al., Evolutionary History and Functional Diversification of Phosphomannomutase Genes[http://www.springerlink.com.ezproxy.library.wisc.edu/content/f677074356610475/fulltext.pdf]</ref>. All of the residues that play the critical role in catalytic activity of the protein are well <scene name='Sandbox89220/2amy/22'>conserved</scene> throughout the evolutionary path. The cap domain of PMM2 is smaller, functioning as the regulator for the access of the substrate into the active site of the core domain and at the same time contains the primary and secondary substrate specificity loops that recognize only specific substrate. It's still unclear how the PMM2 would work together to catalyze the transfer of phosphoryl group within its substrate, but it's possible that it might have the same machinery as the PGM(phosphoglycerate mutase) protein family. | <scene name='Sandbox89220/2amy/12'>PMM2</scene> is a 246 amino acid long protein, with 2 identical subunits having close resemblance in their <scene name='Sandbox89220/2amy/17'>secondary structure</scene> working as independent domains, hence it’s also known as a homodimeric protein. Even if the structure of PMM2 is already solved,however, it's still not clear where its domains and its critical motifs are. Nonetheless, through the extraploration of PMM1, it’s known to be classified as the member of HAD (haloacid dehydrolase) superfamily due to the fact that it had 4 different types of motifs[http://www.sinauer.com/pdf/nsp-protein-1-16.pdf], each of which is highly conserved across the vertebrates’ lineages during evolutions. Similar to other members in HAD superfamily, the two domains are separately recognized as core and cap domain. The core domain (residues 1 – 90, and 198-262)of PMM2 displays the characteristic 4 motifs of HAD superfamily that contribute to the catalytic ability of the active sites in the protein. Motif 1 has <scene name='Sandbox89220/2amy/25'>Asp</scene> as nucleophile which serves as the mediator for the phosphoryl group transfer, the second Asp, on the other hand, acts as in the general acid-base reaction. While motif 1 provides the nucleophilic and general acid-base reactions, motif 2 in the protein which usually contains <scene name='Sandbox89220/2amy/26'>Thr</scene> or <scene name='Sandbox89220/2amy/28'>Ser</scene> helps in positioning or binding on the substrates’ phosphoryl group. In addition, motif 3 has the typical Lys or Arg residues while motif 4 has the acidic residues (Asp and Glu) that bind magnesium cofactor[http://en.wikipedia.org/wiki/Cofactor_%28biochemistry%29]<ref name="Evolutionary history">Rita Quental et al., Evolutionary History and Functional Diversification of Phosphomannomutase Genes[http://www.springerlink.com.ezproxy.library.wisc.edu/content/f677074356610475/fulltext.pdf]</ref>. All of the residues that play the critical role in catalytic activity of the protein are well <scene name='Sandbox89220/2amy/22'>conserved</scene> throughout the evolutionary path. The cap domain of PMM2 is smaller, functioning as the regulator for the access of the substrate into the active site of the core domain and at the same time contains the primary and secondary substrate specificity loops that recognize only specific substrate. It's still unclear how the PMM2 would work together to catalyze the transfer of phosphoryl group within its substrate, but it's possible that it might have the same machinery as the PGM(phosphoglycerate mutase) protein family. | ||
Revision as of 01:39, 29 April 2011
Phosphomannose mutase 2
Background
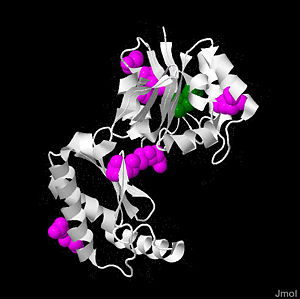
Phosphomannose mutase 2,2amy
Structure
| |||||||||||