User:Momodou L. Jammeh/Sandbox 1
From Proteopedia
| Line 19: | Line 19: | ||
<scene name='User:Momodou_L._Jammeh/Sandbox_1/Rho_p-loop/2'>C-terminus structures</scene> | <scene name='User:Momodou_L._Jammeh/Sandbox_1/Rho_p-loop/2'>C-terminus structures</scene> | ||
| + | |||
| + | <scene name='User:Momodou_L._Jammeh/Sandbox_1/Subunit-c-terminal/1'>C_terminal</scene> | ||
---- | ---- | ||
===MECHANISM=== | ===MECHANISM=== | ||
Revision as of 11:35, 29 April 2011
Contents |
RHO TERMINATION FACTOR
INTRODUCTION
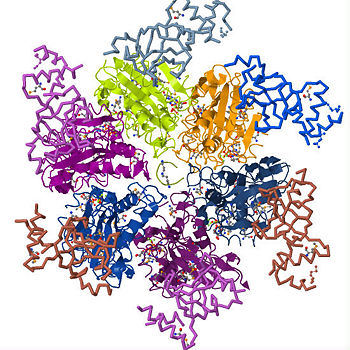
Transcription is the process of mRNA synthesis by RNA polymerase, an enzyme that uses a strand of DNA as a template for ribonucloetide addition. During this process, RNA polymerase encounters several regulatory proteins, called transcription factors, that effect transcription in various ways. One of the major regulatory activities of transcription regulators is terminating the process at specific sites in the transcription unit. In E. coli and prokaryotes in general, transcription is terminated through rho dependent and independent mechanisms. Transcription termination without rho occurs through RNA polymerase's intrinsic ability to terminate transcription in response to certain, limited sequences in the template DNA strand. Rho dependent termination requires a helicase protein called rho factor which belongs in the helicase class of enzymes. Helicases are motor proteins that function through directional movement on the nucleic acid phosphodiester backbone and can dissociate bound DNA-DNA, RNA-DNA or RNA-RNA strands. Rho specifically terminates transcription by separating the the new RNA strand annealed to the DNA strand being transcribed. As pictured on the upper right corner, it is a hexameric ring-shaped helicase that uses energy derived from its ATPase mechanism (hydrolysis of ATP into ADP and an organic phosphate) to drive its movement along the newly formed RNA molecule toward the elongation complex to be dissociated. While rho is only requirement for the termination process, additional elongation factors such as NusA and NusG have been shown to enhance the process. NusA reduces the elongation rate allowing rho to catch up with RNA polymerase despite its high processivity while NusG enhances the rate of RNA release by coupling rho to the elongation complex. Since its discovery in 1969, studies on rho have been essential in understanding how external factors regulation transcription termination and its structure provides insight into how several stable hexameric helicases load onto their nucleic acid substrates.
STRUCTURE
|
Rho is assemble through subunit-subunit interactions between both the amino- and carboxyl terminus domains of the six subunits. Adjacent N-terminal domains make contact through α2/α3 connector loop, while the C-terminus interactions occur through the α11 which packs against the β7/α8 and β8/α9 junctions of the neighboring subunit. Although, extensive reference has been made to existence of rho as a closed ring, it is also thought to split open to yield the "" conformation. An unusual conformation considering the fact that almost all fully crystallized, full length structures of rho are in closed ring form.
MECHANISM
The termination of transcription by rho requires the binding of nascent RNA by the hexamer which functions as an RNA-activated ATPase helicase. This molecular motor is driven by ATP hydrolysis which occurs in the main catalytic site in the C-terminal domain of rho monomers. Rho makes contact with its substrate, nascent RNA transcript, through its OB fold which serves as the primary RNA binding site. The OB fold fold recognizes and binds to the cytosine-rich region of a transcript called the rho utilization (rut) sequence of 40 or more bases. Thus, the N-terminus domain makes first contact with RNA and the loading process begins.