This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
User:Momodou L. Jammeh/Sandbox 1
From Proteopedia
| Line 10: | Line 10: | ||
===Structure=== | ===Structure=== | ||
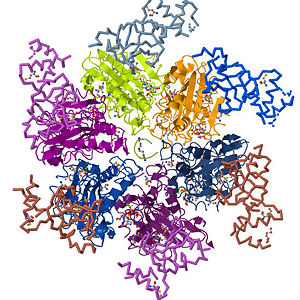
| - | <Structure load='3ICE' size='450' frame='true' align='right' caption='Rho factor in its hexameric form with its six identical subunits forming a helicase and an RNA molecule right through the center' /> Rho factor is a hexamer of six identical subunits that interact to form a closed, dynamic structure called a <scene name='User:Momodou_L._Jammeh/Sandbox_1/Hexameric_rho/2'>helicase.</scene> It is made up of a single polypeptide chain of about 419 amino acid residues which comprise the <scene name='User:Momodou_L._Jammeh/Sandbox_1/Rho_ball_helix/2'>alpha-helix</scene> and <scene name='User:Momodou_L._Jammeh/Sandbox_1/Rho_ball_sheets/3'>beta sheet</scene> conformations that make up the <scene name='User:Momodou_L._Jammeh/Sandbox_1/Rho_ball_secondary/3'>secondary structure</scene> (single subunit). Each subunit has an <scene name='User:Momodou_L._Jammeh/Sandbox_1/Hexameric_rho_n-terminal/3'>amino-terminus</scene>(residues in blue and light blue) and <scene name='User:Momodou_L._Jammeh/Sandbox_1/Hexameric_rho_n-terminal/4'>carboxyl-terminus</scene>(residues in red and orange) domain with the former on the periphery of the subunits and the latter, which is also the main catalytic site on the central, upper portion of the hexamer<ref>Richardson, JP, Structural Organization of Transcription Termination Factor Rho, J. Biol. Chem. 271(1996), pp. 1251 - 1254</ref> . The <scene name='User:Momodou_L._Jammeh/Sandbox_1/Rho_subunit_n-terminal/1'>N-terminal</scene> domain of the rho <scene name='User:Momodou_L._Jammeh/Sandbox_1/Rho_ball_secondary/3'>monomer</scene> (A chain) consists of amino acid residues 1 - 130 and serves as the primary binding site for the C-rich region of the nascent RNA transcript. It has an <scene name='User:Momodou_L._Jammeh/Sandbox_1/Rho_subunit_ob_fold_helix/2'>α-helix (green) capped β-barrel (yellow)</scene> formed from five polypeptide strands that comprise | + | <Structure load='3ICE' size='450' frame='true' align='right' caption='Rho factor in its hexameric form with its six identical subunits forming a helicase and an RNA molecule right through the center' /> Rho factor is a hexamer of six identical subunits that interact to form a closed, dynamic structure called a <scene name='User:Momodou_L._Jammeh/Sandbox_1/Hexameric_rho/2'>helicase.</scene> It is made up of a single polypeptide chain of about 419 amino acid residues which comprise the <scene name='User:Momodou_L._Jammeh/Sandbox_1/Rho_ball_helix/2'>alpha-helix</scene> and <scene name='User:Momodou_L._Jammeh/Sandbox_1/Rho_ball_sheets/3'>beta sheet</scene> conformations that make up the <scene name='User:Momodou_L._Jammeh/Sandbox_1/Rho_ball_secondary/3'>secondary structure</scene> (single subunit). Each subunit has an <scene name='User:Momodou_L._Jammeh/Sandbox_1/Hexameric_rho_n-terminal/3'>amino-terminus</scene>(residues in blue and light blue) and <scene name='User:Momodou_L._Jammeh/Sandbox_1/Hexameric_rho_n-terminal/4'>carboxyl-terminus</scene>(residues in red and orange) domain with the former on the periphery of the subunits and the latter, which is also the main catalytic site on the central, upper portion of the hexamer<ref>Richardson, JP, Structural Organization of Transcription Termination Factor Rho, J. Biol. Chem. 271(1996), pp. 1251 - 1254</ref> . The <scene name='User:Momodou_L._Jammeh/Sandbox_1/Rho_subunit_n-terminal/1'>N-terminal</scene> domain of the rho <scene name='User:Momodou_L._Jammeh/Sandbox_1/Rho_ball_secondary/3'>monomer</scene> (A chain) consists of amino acid residues 1 - 130 and serves as the primary binding site for the C-rich region of the nascent RNA transcript. It has an <scene name='User:Momodou_L._Jammeh/Sandbox_1/Rho_subunit_ob_fold_helix/2'>α-helix (green) capped β-barrel (yellow)</scene> formed from five polypeptide strands that comprise two anti parallel sheets connected by four loops. The barrel is closed by hydrogen bonds between the third and fifth strands and forms the oligonucleotide/oligosaccharide binding (OB) fold of rho. The <scene name='User:Momodou_L._Jammeh/Sandbox_1/Rho_subunit_n-terminal-ob_fold/1'>OB fold</scene> is a folding motif that is common among oligonucleotide or oligosaccharide binding proteins<ref>Hitchens, TK, Zhan, Y, Richardson, LV, Richardson, JP, and Rule, GS, Sequence-specific interactions in the RNA-binding domain of the Escherichia coli transcription termination factor rho, J. Biol. Chem. 2006, 281, pp. 33697 – 33703</ref>. Although OB folds have been found in several other proteins such as staphylococcal nuclease and anti codon binding regions of asp-tRNA synthetase, the geometry of the OB fold (five strand beta-barrel of two antiparallel sheets, otherwise known as the [http://en.wikipedia.org/wiki/Beta_sheet greek key motif]) is the same for all proteins but their is no sequence similarity. Therefore, the binding of these proteins to their oligonucleotide/oligosaccharide substrates depends on fold geometry and topology rather than on the protein sequences<ref>Thomsen, ND, and Berger, JM, Structural frameworks for considering microbial protein- and nucleic acid-dependent motor ATPases, Mol Microbiol 69 (2008), pp. 1071–1090</ref>. Because of the presence of the <scene name='User:Momodou_L._Jammeh/Sandbox_1/Rho_subunit_ob_fold_helix/3'>OB fold</scene> in the N-terminal domain of rho, it is evident that this sub-domain constitutes the primary binding site for the RNA transcript as it loads on to rho. As stated earlier, the <scene name='User:Momodou_L._Jammeh/Sandbox_1/Rho_ball_secondary/4'>C-terminal domain</scene> is the main catalytic site of rho and has several structural motifs to that effect. The major catalytic process that occurs in this domain is the hydrolysis of ATP and thus contains the Walker A and Walker B signature motifs that are common to such ATPases. Here, an <scene name='User:Momodou_L._Jammeh/Sandbox_1/Rho_ball_atpase/4'>ADP molecule</scene> sits in the ATP binding pocket of adjoining A and B subunits of rho. The Walker A motif is the site for ATP binding and contains the <scene name='User:Momodou_L._Jammeh/Sandbox_1/Rho_p-loop-proper/1'>P-loop (in blue)</scene> which has the consensus amino acid sequence GXXXXGK(T/S), with the K denoting the <scene name='User:Momodou_L._Jammeh/Sandbox_1/Rho_p-loop-proper/1'>Lysine residue in yellow</scene> which is crucial for nucleotide binding. In addition to LYS, <scene name='User:Momodou_L._Jammeh/Sandbox_1/Rho_subunit_atpresidues/2'>ARG366(green), ARG212(yellow), THR185(cyan) and PHE355(red)</scene> interact with bound ATP<ref>Burgess, BR, and Richardson, JP, RNA Passes through the Hole of the Protein Hexamer in the Complex with the Escherichia coli Rho Factor, J. Biol. Chem. 276 (2001), pp. 4182 - 4189</ref>. The <scene name='User:Momodou_L._Jammeh/Sandbox_1/Subunits_with_mg2/1'>Mg2+ ion (green)</scene> coordinates the γ- and β-phosphates of nucleotides. The Walker B motif which is characterized by the <scene name='User:Momodou_L._Jammeh/Sandbox_1/Rho_q_and_r_l/1'>Q-loop in yellow</scene> and <scene name='User:Momodou_L._Jammeh/Sandbox_1/Rho_q_and_r_l/1'>R-loop in orange</scene> facilitates mRNA translocation and unwinding and thus comprises the secondary binding site of the <scene name='User:Momodou_L._Jammeh/Sandbox_1/Complete_c-terminus/1'>C-terminus domain</scene>. Rho is assemble through subunit-subunit interactions between both the amino- and carboxyl terminus domains of the six subunits. Adjacent N-terminal domains make contact through α2/α3 connector loop, while the C-terminus interactions occur through the α11 which packs against the β7/α8 and β8/α9 junctions of the neighboring subunit. Although, extensive reference has been made to existence of rho as a closed ring, there is increasing evidence that rho occurs in the "<scene name='User:Momodou_L._Jammeh/Sandbox_1/Rho_washer/1'>lock washer</scene>" conformation when unbound by RNA. This form of rho has been name the "loading" conformation as the gap is wide enough to accommodate the mRNA as it makes its way to the Q-loop<ref>Canals, A, Uson, I, and Coll, M, The Structure of RNA-Free Rho Termination Factor Indicates a Dynamic Mechanism of Transcript Capture. J. Mol. Biol. 2010, 400(1): 16 - 23</ref>. |
---- | ---- | ||
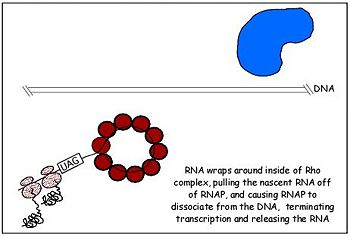
[[Image:rho-4.jpg|350px||left|thumb|Fig. 2 Rho dissociates transcription elongation complex to complete the final step of rho dependent termination]] | [[Image:rho-4.jpg|350px||left|thumb|Fig. 2 Rho dissociates transcription elongation complex to complete the final step of rho dependent termination]] | ||
Revision as of 05:58, 1 May 2011
Contents |
Rho factor
Introduction
Transcription is the process of mRNA synthesis by RNA polymerase, an enzyme that uses a strand of DNA as a template for ribonucloetide addition. During this process, RNA polymerase encounters several regulatory proteins, called transcription factors, that effect transcription in various ways. One of the major regulatory activities of transcription regulators is terminating the process at specific sites in the transcription unit. In E. coli and prokaryotes in general, transcription is terminated through rho dependent and independent mechanisms. Transcription termination without rho occurs through RNA polymerase's intrinsic ability to terminate transcription in response to certain, limited sequences in the template DNA strand. Rho dependent termination requires a helicase protein called rho factor which belongs in the helicase class of enzymes[1][2]. Helicases are motor proteins that function through directional movement on the nucleic acid phosphodiester backbone and can dissociate bound DNA-DNA, RNA-DNA or RNA-RNA complexes. As pictured on the upper right corner, rho is a hexameric ring-shaped helicase that uses energy derived from its ATPase mechanism (hydrolysis of ATP into ADP and an organic phosphate) to drive its movement along the newly formed RNA molecule toward the elongation complex to be dissociated[3]. While rho is the only requirement for the termination process, additional elongation factors such as NusA and NusG have been shown to enhance the process. NusA reduces the elongation rate allowing rho to catch up with RNA polymerase despite its high processivity while NusG enhances the rate of RNA release by coupling rho to the elongation complex. Pause signal sequences are also common at rho termination sites. The elongation complex is halted at these sequences making it more susceptible to rho termination[4] . Since its discovery in 1969, studies on rho have been essential in understanding how external factors regulation transcription termination and its structure provides insight into how several stable hexameric helicases load onto their nucleic acid substrates.
Structure
|
Mechanism
Highlights of 's functional sub-domains. The termination of transcription by rho requires the binding of nascent RNA by the hexamer which functions as an RNA-activated ATPase helicase. This molecular motor is driven by ATP hydrolysis which occurs in the main catalytic site in the C-terminal domain of rho monomers. Rho makes contact with its substrate, nascent RNA transcript, through its OB fold which serves as the primary RNA binding site. The OB fold fold recognizes and binds to the cytosine-rich region of a transcript called the rho utilization (rut) sequence of 40 or more bases. Thus, the N-terminus domain makes first contact with RNA and the loading process begins[10]. Once the OB fold makes contact with the mRNA, a notch forms in rho turning it into an open ring and allows the mRNA to load unto the protein and associate with the Q and R loops of the secondary binding site. The first cytidine base fits tightly within a hydrophobic pocket formed by Phe62 and Tyr80, and it also contacts residues Glu108, Arg109, and Tyr110 in the RNA-binding domain. The second cytidine forms specific hydrogen bonds with Arg66 and Asp78 and stacks on the aromatic side chain of Phe64. This stimulates ring closer and also alters the conformation of the P-loop such that it is appropriate for ATP binding and subsequent hydrolysis, an ATPase. Although rho has six potential ATPase sites, it functions as a trimer of dimers through sequential firing of three of the ATPase sites. Rho's ATPase activity is stimulated by the mRNA whose fourth nucloetide from the 5' end is bound by Lys181 and orients it toward Lys184 which links the ATPase active site to the RNA substrate. The energy derived from ATP hydrolysis drives the translocation of rho as it moves along the nucleic acid track. This second step in rho dependent termination occurs in 5' to 3' fashion as the RNA threads through the central cavity of rho. In addition, the RNA also wraps around the top part of the low affinity c-terminal residues of rho as it moves toward the RNA polymer-DNA complex which it eventually dissociates with forces from ATP hydrolysis[11].
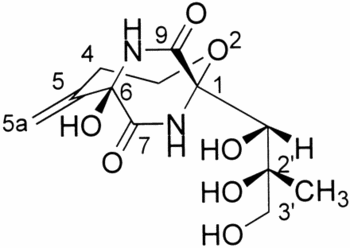
Bicyclomycin
|
References
- ↑ Singleton, MR, Dillingham, MS, and Wigley, DB, Structure and mechanism of helicases and nucleic acid translocases, Annu. Rev. Biochem. 76 (2007), pp. 23 - 50
- ↑ Banerjee S, Chalissery J, Bandey I, Sen R: Rho-dependent transcription termination: more questions than answers. J Microbiol 2006, 44:11-22
- ↑ Stitt, BL, Xu, Y, Sequential Hydrolysis of ATP Molecules Bound in Interacting Catalytic Sites of Escherichia coli Transcription Termination Protein Rho, J. Biol. Chem. 1998 273 (41) 26477-26486
- ↑ Ingham, CJ, Dennis, J, and Furneaux, PA, Autogenous regulation of transcription termination factor Rho and the requirement for Nus factors in Bacillus subtilis, Mol. Microbiology, 31 (1999), 651 – 663
- ↑ Richardson, JP, Structural Organization of Transcription Termination Factor Rho, J. Biol. Chem. 271(1996), pp. 1251 - 1254
- ↑ Hitchens, TK, Zhan, Y, Richardson, LV, Richardson, JP, and Rule, GS, Sequence-specific interactions in the RNA-binding domain of the Escherichia coli transcription termination factor rho, J. Biol. Chem. 2006, 281, pp. 33697 – 33703
- ↑ Thomsen, ND, and Berger, JM, Structural frameworks for considering microbial protein- and nucleic acid-dependent motor ATPases, Mol Microbiol 69 (2008), pp. 1071–1090
- ↑ Burgess, BR, and Richardson, JP, RNA Passes through the Hole of the Protein Hexamer in the Complex with the Escherichia coli Rho Factor, J. Biol. Chem. 276 (2001), pp. 4182 - 4189
- ↑ Canals, A, Uson, I, and Coll, M, The Structure of RNA-Free Rho Termination Factor Indicates a Dynamic Mechanism of Transcript Capture. J. Mol. Biol. 2010, 400(1): 16 - 23
- ↑ Richardson, JP, Loading Rho to terminate Transcription, Cell, 114 (2003), pp. 157 – 159
- ↑ Skordalakes, E, and Berger,JM, Structural insights into RNA-dependent ring closure and ATPase activation by the Rho termination factor, Cell 127 (2006), pp. 553–564
- ↑ Skordalakes, E, Brogen, AP, Park, BS, Kohn, H, and Berger, JM, Structural Mechanism of Inhibition of the Rho Transcription Termination Factor by the Antibiotic Bicyclomycin, Structure, 13(2005):1, pp. 99 - 109
- ↑ Zwiefka, A, Kohn, H, Transcription termination factor rho: The site of bicyclomycin inhibition in Escherichia coli, Biochemistry, 1993, 32 (14), pp. 3564 - 3570