DOPA decarboxylase
From Proteopedia
| Line 16: | Line 16: | ||
---- | ---- | ||
===The Active Site=== | ===The Active Site=== | ||
| - | The active site of DOPA decarboxylase is located in a cleft at the <scene name='DOPA_decarboxylase/Dimer_interface/2'>interface</scene> between the two subunits of the dimer, like all PLP-dependent enzymes of the aspartate aminotransferase family. Since it is at the interface, residues from both domains and both subunits are involved in cofactor binding, although the active site is composed of residues mainly from one monomer. The <scene name='DOPA_decarboxylase/Active_site/1'>active site</scene> is composed of several key residues. <scene name='DOPA_decarboxylase/Lysine2/1'>Lys303</scene> serves to bind PLP via a [http://en.wikipedia.org/wiki/Schiff_base Schiff base] linkage. As well, a salt bridge exists between the carboxyl group of <scene name='DOPA_decarboxylase/ | + | The active site of DOPA decarboxylase is located in a cleft at the <scene name='DOPA_decarboxylase/Dimer_interface/2'>interface</scene> between the two subunits of the dimer, like all PLP-dependent enzymes of the aspartate aminotransferase family. Since it is at the interface, residues from both domains and both subunits are involved in cofactor binding, although the active site is composed of residues mainly from one monomer. The <scene name='DOPA_decarboxylase/Active_site/1'>active site</scene> is composed of several key residues. <scene name='DOPA_decarboxylase/Lysine2/1'>Lys303</scene> serves to bind PLP via a [http://en.wikipedia.org/wiki/Schiff_base Schiff base] linkage. As well, a salt bridge exists between the carboxyl group of <scene name='DOPA_decarboxylase/Aspartic/1'>Asp271</scene> and a pyridine nitrogen of PLP to further stabilize intermediates. |
The <scene name='Sandbox/Active_site3/1'>inhibitor</scene> binds to the enzyme by forming a hydrazone linkage | The <scene name='Sandbox/Active_site3/1'>inhibitor</scene> binds to the enzyme by forming a hydrazone linkage | ||
==Classification== | ==Classification== | ||
Revision as of 04:01, 10 May 2011
Contents |
Introduction
| |||||||||
| 1js3, resolution 2.25Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , , | ||||||||
| Activity: | Aromatic-L-amino-acid decarboxylase, with EC number 4.1.1.28 | ||||||||
| Related: | 1js6 | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
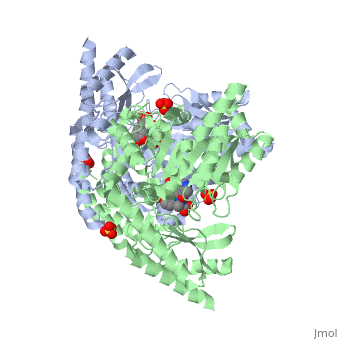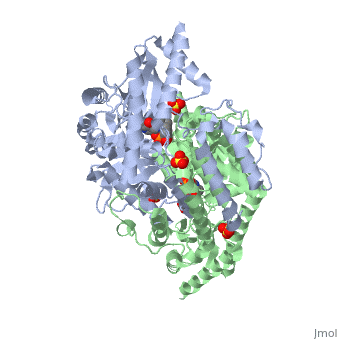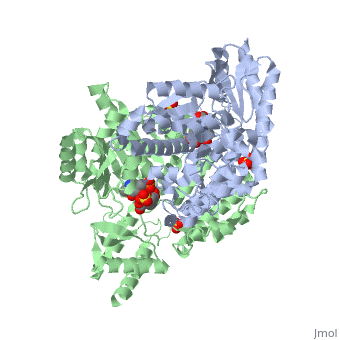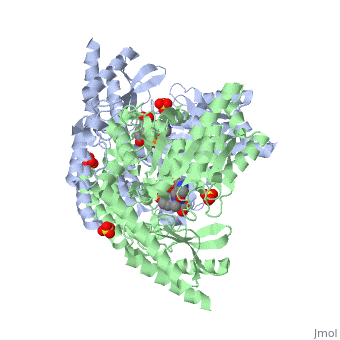
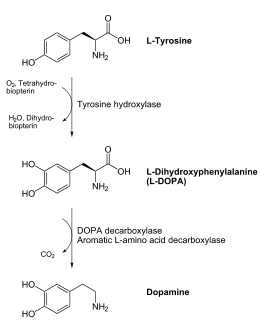
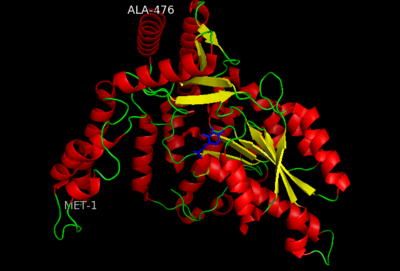
DOPA decarboxylase (DDC, aromatic L-amino acid decarboxylase, tryptophan decarboxylase, 5-hydroxytryptophan decarboxylase, AAAD) is an approximately 104 kDa protein that belongs to the aspartate aminotransferase family (fold type 1) of PLP-dependent enzymes. The catalytically active form of the enzyme exists as a homodimer, typical of this class of enzymes.[1] The homodimeric form of the enzyme purified from sus scrofa is shown in complex with the inhibitor carbidopa to the right. DOPA decarboxylase is responsible for the synthesis of dopamine and serotonin from L-DOPA and L-5-hydroxytryptophan, respectively. Due to its role in neurotransmitter synthesis, DOPA decarboxylase has been implicated in Parkinson's disease, a disease thought to be the result of the degeneration of dopamine-producing cells in the brain. Currently, treatment for the disease is aimed at DOPA decarboxylase inhibition, which would allow greater amounts of exogenously administered L-DOPA to reach the brain. [2]
Structure
The Aspartate Aminotransferase Family
PLP-dependent enzymes belonging to this family are catalytically active as homodimers and share a common, well-characterized structure. Each subunit has a large domain and a small domain. The central feature of the large domain is a seven-stranded β sheet. The small domain has either a three or four-stranded β sheet that is surrounded by helices on one side. The cofactor PLP is covalently attached to a lysine residue in the large domain and is anchored in a way that allows the aromatic ring of PLP to pack against neighboring β strands.
DOPA decarboxylase
DOPA decarboxylase is a homodimeric enzyme, with each monomer composed of three distinct domains. The large domain contains the PLP binding site, and consists of a seven stranded mixed beta-sheet that is surrounded by eight alpha-helices, resulting in a typical α/β fold. The C-terminal domain is comprised of a four-stranded anti-parallel beta-sheet that has three alpha-helices packed against the face opposite to the large domain. Although the aforementioned domains exist in all members of this family of PLP-dependent enzymes, including bacterial ornithine decarboxylase (OrnDC) and dialkylglycine decarboxylase (DGD), the is unique to DOPA decarboxylase, and is a representative case of domain swapping. This domain is composed of two parallel helices linked by an extended strand, which essentially lies like a flap over the second subunit. The N-terminal domain of one monomer packs on top of the other monomer, resulting in an extended dimer interface, and thus it is most likely stable only in the dimeric form of the enzyme.
Function
The Active Site
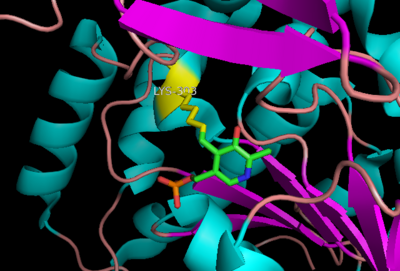
The active site of DOPA decarboxylase is located in a cleft at the between the two subunits of the dimer, like all PLP-dependent enzymes of the aspartate aminotransferase family. Since it is at the interface, residues from both domains and both subunits are involved in cofactor binding, although the active site is composed of residues mainly from one monomer. The is composed of several key residues. serves to bind PLP via a Schiff base linkage. As well, a salt bridge exists between the carboxyl group of and a pyridine nitrogen of PLP to further stabilize intermediates. The binds to the enzyme by forming a hydrazone linkage
Classification
SCOP
DOPA decarboxylase is classified in the following manner using SCOP:
- Class: alpha and beta proteins (α/β)
- Fold: PLP-dependent transferase-like
- Superfamily: PLP-dependent transferases
- Family: Pyridoxal-dependent decarboxylase
- Domain""": DOPA decarboxylase
CATH
DOPA decarboxylase is classified in the following manner using CATH:
- large domain
- Class: alpha beta
- Architecture: 3-layer sandwich
- Topology: Aspartate aminotransferase
- small domain
- Class: alpha beta
- Architecture: alpha-beta complex
- Topology: Aspartate aminotransferase
- N-terminal domain
- Class: mainly alpha
- Architecture: up-down bundle
- Topology: dopa decarboxylase
References
- ↑ Schneider G, Kack H, Lindqvist Y. The manifold of vitamin B6 dependent enzymes. Structure. 2000 Jan 15;8(1):R1-6. PMID:10673430
- ↑ Burkhard P, Dominici P, Borri-Voltattorni C, Jansonius JN, Malashkevich VN. Structural insight into Parkinson's disease treatment from drug-inhibited DOPA decarboxylase. Nat Struct Biol. 2001 Nov;8(11):963-7. PMID:11685243 doi:http://dx.doi.org/10.1038/nsb1101-963
Proteopedia Page Contributors and Editors (what is this?)
Brittany Todd, Michal Harel, David Canner, Alexander Berchansky, Brian Hernandez