This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
User:Wayne Decatur/kink-turn motif
From Proteopedia
m (added schemetic) |
m |
||
| Line 19: | Line 19: | ||
{{Button Toggle NucleicDrumsColorScheme}} {{ColorKey Bases RNA}} | {{Button Toggle NucleicDrumsColorScheme}} {{ColorKey Bases RNA}} | ||
</center></td></tr></table> | </center></td></tr></table> | ||
| - | Originally identified in the course of analyzing the [[Large Ribosomal Subunit of Haloarcula|large ribosomal subunit]]<ref name="kleinref">PMID: 11483524</ref>, this RNA structural motif was also identified in other RNAs. Particular instances have been called the GA motif <ref>PMID: 11497434</ref> | + | [[kink-turn motif|The kink-turn motif]] is a common RNA structural motif that consists of helix–internal loop–helix motif that introduces a very tight kink into the helical axis. Originally identified in the course of analyzing the [[Large Ribosomal Subunit of Haloarcula|large ribosomal subunit]]<ref name="kleinref">PMID: 11483524</ref>, this RNA structural motif was also identified in other RNAs. Particular instances have been called the GA motif <ref>PMID: 11497434</ref>. |
The kink-turn motif includes the [[A-minor motif]]. | The kink-turn motif includes the [[A-minor motif]]. | ||
Many kink-turns bind proteins; however, that trait is not universal. They can mediate RNA tertiary structure interactions as well. | Many kink-turns bind proteins; however, that trait is not universal. They can mediate RNA tertiary structure interactions as well. | ||
Revision as of 22:07, 15 May 2011
| For the date when the most recent work on this article was done, click on the history tab above. |
WHEN MADE INTO AN OFFICIAL PAGE:
- LINK FROM RNA motifs PAGE!!!!
- Fix links from and to large ribosomal subunit (AND RIBOSOME?) page
- Fix link from topic pages and from table of contents
- add Redirect from K-turn motif
- add redirect from GA motif
- add redirect from kink-turn
- add redirect from k-turn
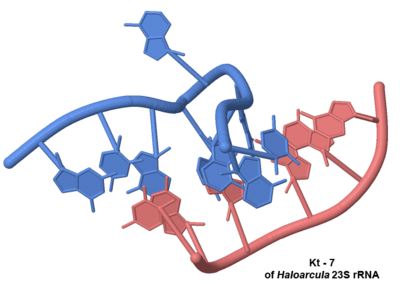
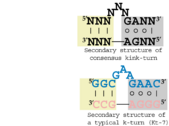
The kink-turn motif
A common RNA structural motif that consists of helix–internal loop–helix motif that introduces a very tight kink into the helical axis.
Contents |
Introduction
| |||||||
A C G U |
The kink-turn motif is a common RNA structural motif that consists of helix–internal loop–helix motif that introduces a very tight kink into the helical axis. Originally identified in the course of analyzing the large ribosomal subunit[1], this RNA structural motif was also identified in other RNAs. Particular instances have been called the GA motif [2]. The kink-turn motif includes the A-minor motif. Many kink-turns bind proteins; however, that trait is not universal. They can mediate RNA tertiary structure interactions as well.
An excellent introduction can be found here as part of a structural database for k-turn motifs in RNA by the Lilley group[3].
Structures Containing the Motif
| |||||||
A C G U |
- The Large Ribosomal Subunit contains 9 identified kink-turns[1][3]: and Kt-7 ; and Kt-15 ; ; and Kt-42 ; and Kt-46 ); and Kt-58 ; and Kt-4/5 ; and Kt-94/99 with of 3cc2; as observed in 2wh2 (While present based on sequence comparison, Kt-78 is in a portion of the large subunit not visible in the crystal structure of 3cc2.)
- observed in the solved structure of the Haloarcula large ribosomal subunit shows they are on the surface of the subunit: , , , , , , , and . (A ninth kink-turn, Kt-78, though present in Haloarcula marismortui rRNA is in a portion not resolved in the actual Haloarcula marismortui structure, yet is also on the surface.)
- See all of the 23S rRNA kink-turns and others on this page in greater detail following the appropriate links on this page at a structural database for k-turn motifs in RNA by the Lilley group[3].
- The human spliceosomal and small nucleolar RNA-binding 15.5kD protein bound to the kink-turn of a U4 spliceosomal RNA fragment (1e7k)[4]. The fact both the box C/D small nucleolar RNAs and the spliceosomal U4 RNA share this motif, and in fact bind the same protein, was observed[5][4] even before the kink-turn was shown to be a widespread RNA motif.
- The from Sulfolobus solfataricus has two kink-turns that are bound by L7Ae (3pla)[6]. .
- A. fulgidus small ribonucleoprotein particle box C/D RNA has a kink-turn that is bound by L7Ae (1rlg)[7] has a kink turn.
- Pyrococcus furiosus small ribonucleoprotein particle box H/ACA RNA (2hvy[8],3hax,3hay[9]) has a kink turn that is bound directly by L7ae in a complex of several proteins.
- Azoarcus group I intron (1u6b, 1zzn, 3bo2, 3bo3, 3bo4, and 3iin)[10][11][12][13] has a 'reverse' kink-turn. Overlay of the Azoarcus group I intron reverse kink-turn with a typical one (Kt-7) clearly illustrates the difference. PUT A SCENE HERE OF ALIGNMENT OF THIS WITH KT-7 with each Kt colored differently
- S. cervisiae L30e bound to its pre-mRNA (1t0k)[14] has a kink-turn with a protein bound.
- S-adenosylmethionine riboswitch regulatory mRNA element from Thermoanaerobacter tengcongensis (2gis)[15] has a kink-turn.
- The lysine riboswitch regulatory mRNA element from Thermotoga maritima (3dox) has a kink-turn
- 1nkw – The Large Ribosomal Subunit From Deinococcus radiodurans[16]
- The small ribosomal subunit (2wh1) has two kink-turns.
See Also
- RNA motifs
- Ribosome
- 1go1, 1go0, 1w3e and 1h7m – the Thermococcus celer ribosomal protein L30[17][18]
- A-minor motif
- The adenosine wedge motif[19]
- The G-ribo motif[20]
- The lonepair triloop motif[21]
- RNA ribose zipper[22]
References
- ↑ 1.0 1.1 Klein DJ, Schmeing TM, Moore PB, Steitz TA. The kink-turn: a new RNA secondary structure motif. EMBO J. 2001 Aug 1;20(15):4214-21. PMID:11483524 doi:http://dx.doi.org/10.1093/emboj/20.15.4214
- ↑ Winkler WC, Grundy FJ, Murphy BA, Henkin TM. The GA motif: an RNA element common to bacterial antitermination systems, rRNA, and eukaryotic RNAs. RNA. 2001 Aug;7(8):1165-72. PMID:11497434
- ↑ 3.0 3.1 3.2 Schroeder KT, McPhee SA, Ouellet J, Lilley DM. A structural database for k-turn motifs in RNA. RNA. 2010 Aug;16(8):1463-8. Epub 2010 Jun 18. PMID:20562215 doi:10.1261/rna.2207910
- ↑ 4.0 4.1 Vidovic I, Nottrott S, Hartmuth K, Luhrmann R, Ficner R. Crystal structure of the spliceosomal 15.5kD protein bound to a U4 snRNA fragment. Mol Cell. 2000 Dec;6(6):1331-42. PMID:11163207
- ↑ Watkins NJ, Segault V, Charpentier B, Nottrott S, Fabrizio P, Bachi A, Wilm M, Rosbash M, Branlant C, Luhrmann R. A common core RNP structure shared between the small nucleoar box C/D RNPs and the spliceosomal U4 snRNP. Cell. 2000 Oct 27;103(3):457-66. PMID:11081632
- ↑ Lin J, Lai S, Jia R, Xu A, Zhang L, Lu J, Ye K. Structural basis for site-specific ribose methylation by box C/D RNA protein complexes. Nature. 2011 Jan 27;469(7331):559-563. PMID:21270896 doi:10.1038/nature09688
- ↑ Moore T, Zhang Y, Fenley MO, Li H. Molecular basis of box C/D RNA-protein interactions; cocrystal structure of archaeal L7Ae and a box C/D RNA. Structure. 2004 May;12(5):807-18. PMID:15130473 doi:http://dx.doi.org/10.1016/j.str.2004.02.033
- ↑ Li L, Ye K. Crystal structure of an H/ACA box ribonucleoprotein particle. Nature. 2006 Sep 21;443(7109):302-7. Epub 2006 Aug 30. PMID:16943774 doi:http://dx.doi.org/10.1038/nature05151
- ↑ Duan J, Li L, Lu J, Wang W, Ye K. Structural mechanism of substrate RNA recruitment in H/ACA RNA-guided pseudouridine synthase. Mol Cell. 2009 May 14;34(4):427-39. PMID:19481523 doi:10.1016/j.molcel.2009.05.005
- ↑ Adams PL, Stahley MR, Kosek AB, Wang J, Strobel SA. Crystal structure of a self-splicing group I intron with both exons. Nature. 2004 Jul 1;430(6995):45-50. Epub 2004 Jun 2. PMID:15175762 doi:10.1038/nature02642
- ↑ Stahley MR, Strobel SA. Structural evidence for a two-metal-ion mechanism of group I intron splicing. Science. 2005 Sep 2;309(5740):1587-90. PMID:16141079 doi:309/5740/1587
- ↑ Lipchock SV, Strobel SA. A relaxed active site after exon ligation by the group I intron. Proc Natl Acad Sci U S A. 2008 Apr 15;105(15):5699-704. Epub 2008 Apr 11. PMID:18408159
- ↑ Antonioli AH, Cochrane JC, Lipchock SV, Strobel SA. Plasticity of the RNA kink turn structural motif. RNA. 2010 Apr;16(4):762-8. Epub 2010 Feb 9. PMID:20145044 doi:10.1261/rna.1883810
- ↑ Chao JA, Williamson JR. Joint X-ray and NMR refinement of the yeast L30e-mRNA complex. Structure. 2004 Jul;12(7):1165-76. PMID:15242593 doi:10.1016/j.str.2004.04.023
- ↑ Montange RK, Batey RT. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature. 2006 Jun 29;441(7097):1172-5. PMID:16810258 doi:10.1038/nature04819
- ↑ Harms J, Schluenzen F, Zarivach R, Bashan A, Gat S, Agmon I, Bartels H, Franceschi F, Yonath A. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell. 2001 Nov 30;107(5):679-88. PMID:11733066
- ↑ Chen YW, Bycroft M, Wong KB. Crystal structure of ribosomal protein L30e from the extreme thermophile Thermococcus celer: thermal stability and RNA binding. Biochemistry. 2003 Mar 18;42(10):2857-65. PMID:12627951 doi:10.1021/bi027131s
- ↑ Wong KB, Lee CF, Chan SH, Leung TY, Chen YW, Bycroft M. Solution structure and thermal stability of ribosomal protein L30e from hyperthermophilic archaeon Thermococcus celer. Protein Sci. 2003 Jul;12(7):1483-95. PMID:12824494 doi:10.1110/ps.0302303
- ↑ Gagnon MG, Steinberg SV. The adenosine wedge: a new structural motif in ribosomal RNA. RNA. 2010 Feb;16(2):375-81. Epub 2009 Dec 28. PMID:20038632 doi:10.1261/rna.1550310
- ↑ Steinberg SV, Boutorine YI. G-ribo: a new structural motif in ribosomal RNA. RNA. 2007 Apr;13(4):549-54. Epub 2007 Feb 5. PMID:17283211 doi:10.1261/rna.387107
- ↑ Lee JC, Cannone JJ, Gutell RR. The lonepair triloop: a new motif in RNA structure. J Mol Biol. 2003 Jan 3;325(1):65-83. PMID:12473452
- ↑ Tamura M, Holbrook SR. Sequence and structural conservation in RNA ribose zippers. J Mol Biol. 2002 Jul 12;320(3):455-74. PMID:12096903
Additional Literature and External Resources
- Kink-turns in RNA - a structural database for k-turn motifs in RNA by the Lilley group[1]
- Schroeder KT, McPhee SA, Ouellet J, Lilley DM. A structural database for k-turn motifs in RNA. RNA. 2010 Aug;16(8):1463-8. Epub 2010 Jun 18. PMID:20562215 doi:10.1261/rna.2207910
- Tiedge H. K-turn motifs in spatial RNA coding. RNA Biol. 2006 Oct;3(4):133-9. Epub 2006 Oct 31. PMID:17172877
Categories: RNA | Haloarcula marismortui | Protein-rna | Metal-binding | Ribosome assembly | Tunnel | Ribozyme | Rna-rna | 50S | Large ribosomal subunit | Ribosome | Translation | Kink-turn | K-turn | GA motif | A-minor motif | Ribose zipper | Rna-binding | Rrna-binding | Riboswitch | Topic Page