User:Cameron Ball/Sandbox 1
From Proteopedia
| Line 1: | Line 1: | ||
Replication Terminator Protein (RTP) is a protein found in ''Bacillus Subtilis''(''B.Subtilis'') that plays an important role in the termination of bacterial chromosome replication. RTP binds to the circular bacterial genome to block the progression of DNA polymerase in a polar manner. A homologue of RTP have been found in ''Eschericia coli'' (''E.coli''), named “Termination Utilisation Substance” (Tus) | Replication Terminator Protein (RTP) is a protein found in ''Bacillus Subtilis''(''B.Subtilis'') that plays an important role in the termination of bacterial chromosome replication. RTP binds to the circular bacterial genome to block the progression of DNA polymerase in a polar manner. A homologue of RTP have been found in ''Eschericia coli'' (''E.coli''), named “Termination Utilisation Substance” (Tus) | ||
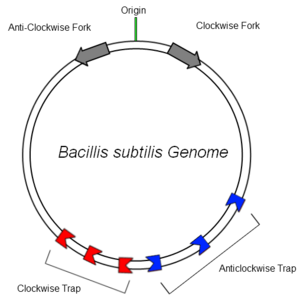
| - | [[Image:bsubtilisrtpschematic.png|300px| | + | [[Image:bsubtilisrtpschematic.png|300px|right|thumb| Schematic diagram of the ''B.subtilis'' genome showing clockwise and anticlockwise replication forks and traps. RTP is represented by the red and blue blocks.]] |
==Introduction== | ==Introduction== | ||
| - | The bacterial genome of ''B.subtilis'' is circular and contains only one origin of replication (OriC). In order to increase the efficiency of DNA replication, the DNA is copied in clockwise and anticlockwise directions simultaneously and later ligated together. It has been found that many organisms employ a mechanism to aid in this termination, suggesting an evolutionary advantage in possessing such a system | + | The bacterial genome of ''B.subtilis'' is circular and contains only one origin of replication (OriC). In order to increase the efficiency of DNA replication, the DNA is copied in clockwise and anticlockwise directions simultaneously and later ligated together. It has been found that many organisms employ a mechanism to aid in this termination, suggesting an evolutionary advantage in possessing such a system <ref>A.A. Griffiths, P.A. Andersen and R.G. Wake, Replication terminator protein-based replication fork-arrest systems in various Bacillus species, J. Bacteriol. 180 (1998), pp. 3360–3367</ref> |
==The Termination Sites== | ==The Termination Sites== | ||
| - | RTP binds DNA as a dimer at Ter sites. These Ter sites are 29 base pairs in length | + | RTP binds DNA as a dimer at Ter sites. These Ter sites are 29 base pairs in length comprise of two non-identical inverted repeats that share three highly conserved base pairs. The two sites are designated the A-site and the B-site and each have different affinities for RTP. It is now known that RTP binds to Ter site in a directional manner as a result of these non-identical sites, allowing polymerase units to pass by one way but blocking them in another. However, historically, the exact way in which symmetric RTP dimers blocked polymerases in a polar fashion was a source of great confusion. This was compounded by the fact that a symmetrical DNA sequence (designated sRB) was used in the first crystal structure of an <scene name='User:Cameron_Ball/Sandbox_1/Rtp_dna_complex/1'>RTP:DNA complex</scene>. <ref>Wilce, J. A., Vivian, J. P., Hastings, A. F., Otting, G., Folmer, R. H., Duggin, I. G., Wake, R. G. & Wilce, M. C., Structure of the RTP-DNA complex and the mechanism of polar replication fork arrest., (2001). Nature Struct. Biol.8, 206–210.</ref> |
| - | ''B.subtilis'' has multiple Ter sites of both polarities to ensure that replication is terminated. The sites are situated off center in relation to the OriC to provide some redundancy at the recombination site | + | ''B.subtilis'' has multiple Ter sites of both polarities to ensure that replication is terminated. The sites are situated off center in relation to the OriC to provide some redundancy at the recombination site. This ensures the entire genome is copied faithfully. |
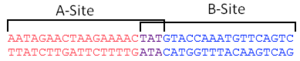
| - | + | [[Image:Ter_site_DNA.png|300px|left|thumb| DNA sequence of a Ter site showing the A-site in Red, the B-site in Blue and the 3 base pair overlap in purple]] | |
| + | |||
| + | == Structure & Mechanism== | ||
| + | <Structure load='1f4k' size='300' frame='true' align='right' caption='RTP bound to symmetric DNA oligonucleotide (sRB)' scene='Insert optional scene name here' /> | ||
| + | <scene name='User:Cameron_Ball/Sandbox_1/Rtp_dna_complex/1'>RTP binds DNA</scene> through interactions between the <scene name='User:Cameron_Ball/Sandbox_1/Rtp_alpha_helices_and_dna_/1'>alpha helices</scene> and the major groove of DNA. Two possible mechanisms were originally proposed to explain it's activity; The "clamp" model, where RTP physically blocks the approaching polymerase and the "interaction" model, where there is some protein-protein interaction between RTP and the approaching replisome unit. Recent data shows that RTP-Ter contact is not sufficient to cause arrest, suggesting the interaction model is the best approximation. <ref> Kaplan, D.L and Bastia, D, Mechanisms of polar arrest of a replication fork, Molecular Microbiology 2009|72|2), 279-285 </ref> For example, in 2006 Ian Duggin showed that the fork arrest efficiency of RTP can be lowered by attaching peptides that would block any protein-protein interaction from occurring while not affecting the RTP-DNA binding affinity<ref> Duggin, I.G., DNA replication fork arrest by the Bacillus subtilis RTP-DNA complex involves a mechanism that is independent of the affinity of RTP-DNA binding ''J Mol Biol'' '''361:'''1-6</ref>. | ||
| - | == | + | ==References== |
| - | + | {{Reflist}} | |
| - | + | ||
Revision as of 08:23, 20 May 2011
Replication Terminator Protein (RTP) is a protein found in Bacillus Subtilis(B.Subtilis) that plays an important role in the termination of bacterial chromosome replication. RTP binds to the circular bacterial genome to block the progression of DNA polymerase in a polar manner. A homologue of RTP have been found in Eschericia coli (E.coli), named “Termination Utilisation Substance” (Tus)
Contents |
Introduction
The bacterial genome of B.subtilis is circular and contains only one origin of replication (OriC). In order to increase the efficiency of DNA replication, the DNA is copied in clockwise and anticlockwise directions simultaneously and later ligated together. It has been found that many organisms employ a mechanism to aid in this termination, suggesting an evolutionary advantage in possessing such a system [1]
The Termination Sites
RTP binds DNA as a dimer at Ter sites. These Ter sites are 29 base pairs in length comprise of two non-identical inverted repeats that share three highly conserved base pairs. The two sites are designated the A-site and the B-site and each have different affinities for RTP. It is now known that RTP binds to Ter site in a directional manner as a result of these non-identical sites, allowing polymerase units to pass by one way but blocking them in another. However, historically, the exact way in which symmetric RTP dimers blocked polymerases in a polar fashion was a source of great confusion. This was compounded by the fact that a symmetrical DNA sequence (designated sRB) was used in the first crystal structure of an . [2]
B.subtilis has multiple Ter sites of both polarities to ensure that replication is terminated. The sites are situated off center in relation to the OriC to provide some redundancy at the recombination site. This ensures the entire genome is copied faithfully.
Structure & Mechanism
|
through interactions between the and the major groove of DNA. Two possible mechanisms were originally proposed to explain it's activity; The "clamp" model, where RTP physically blocks the approaching polymerase and the "interaction" model, where there is some protein-protein interaction between RTP and the approaching replisome unit. Recent data shows that RTP-Ter contact is not sufficient to cause arrest, suggesting the interaction model is the best approximation. [3] For example, in 2006 Ian Duggin showed that the fork arrest efficiency of RTP can be lowered by attaching peptides that would block any protein-protein interaction from occurring while not affecting the RTP-DNA binding affinity[4].
References
- ↑ A.A. Griffiths, P.A. Andersen and R.G. Wake, Replication terminator protein-based replication fork-arrest systems in various Bacillus species, J. Bacteriol. 180 (1998), pp. 3360–3367
- ↑ Wilce, J. A., Vivian, J. P., Hastings, A. F., Otting, G., Folmer, R. H., Duggin, I. G., Wake, R. G. & Wilce, M. C., Structure of the RTP-DNA complex and the mechanism of polar replication fork arrest., (2001). Nature Struct. Biol.8, 206–210.
- ↑ Kaplan, D.L and Bastia, D, Mechanisms of polar arrest of a replication fork, Molecular Microbiology 2009|72|2), 279-285
- ↑ Duggin, I.G., DNA replication fork arrest by the Bacillus subtilis RTP-DNA complex involves a mechanism that is independent of the affinity of RTP-DNA binding J Mol Biol 361:1-6